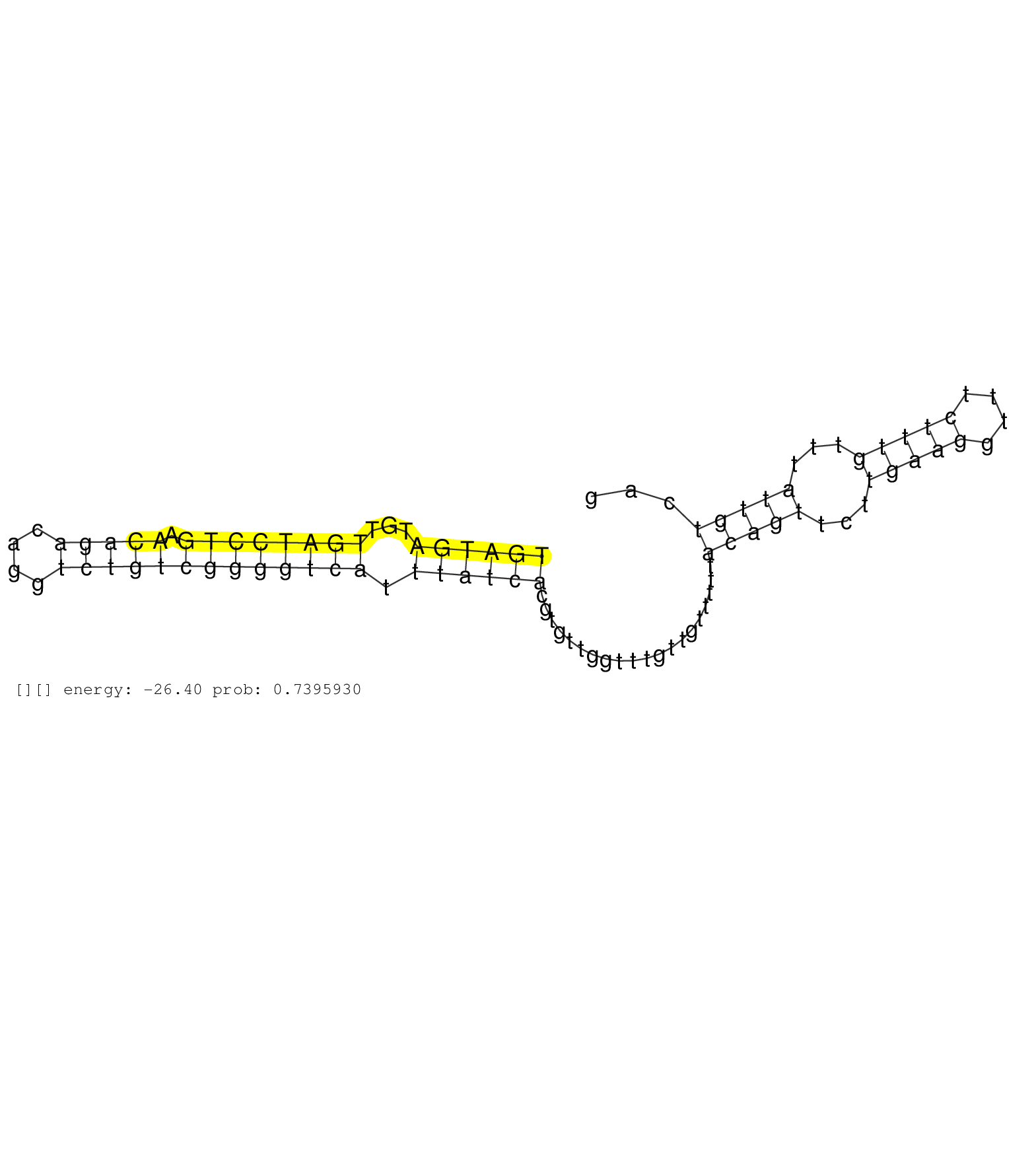
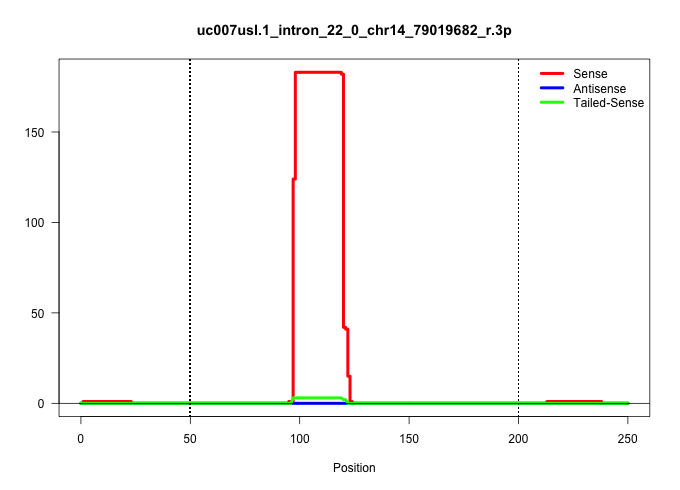
| Gene: Dgkh | ID: uc007usl.1_intron_22_0_chr14_79019682_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| GAGTAGACTGCTCTGGGCAGAGTCACTCACATACAGTGCAAGCTTATTTCCCAGAAGGGTAAATACACCTTCAGTGGTTTCCTAAACACTGAACATTTTGTGATGATGTTGATCCTGAACAGACAGGTCTGTCGGGGTCATTTATCACGTGTTGGTTTGTTGTTTTTACAGTTCTTGAAGGTTTCTTTGTTTATTGTCAGGTGCACACTGCCTGCAAAGATGTGTACCATCCCGTCTGTCCCCTTGGCCA ....................................................................................................((((((...((((((((.(((((....))))))))))))).))))))....................(((((...(((((....)))))...)))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TTGTGATGATGTTGATCCTGAAC.................................................................................................................................. | 23 | 1 | 91.00 | 91.00 | 33.00 | 32.00 | 10.00 | 8.00 | 4.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - |
| ..................................................................................................TGTGATGATGTTGATCCTGAAC.................................................................................................................................. | 22 | 1 | 49.00 | 49.00 | 18.00 | 17.00 | 4.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - |
| .................................................................................................TTGTGATGATGTTGATCCTGAACAG................................................................................................................................ | 25 | 1 | 20.00 | 20.00 | 8.00 | 5.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - |
| .................................................................................................TTGTGATGATGTTGATCCTGAACAGA............................................................................................................................... | 26 | 1 | 16.00 | 16.00 | 6.00 | 1.00 | - | - | 3.00 | 2.00 | 3.00 | - | - | - | - | 1.00 | - |
| ..................................................................................................TGTGATGATGTTGATCCTGAACAG................................................................................................................................ | 24 | 1 | 8.00 | 8.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................TTGTGATGATGTTGATCCTGAACAa................................................................................................................................ | 25 | a | 2.00 | 0.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................TTTGTGATGATGTTGATCCTGAACAGA............................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ..................................................................................................TGTGATGATGTTGATCCTGAACAGA............................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGTGATGATGTTGATCCTGAACA................................................................................................................................. | 23 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................TTTTGTGATGATGTTGATCCTGAAC.................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GCAAAGATGTGTACCATCCCGTCTG............ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................TTGTGATGATGTTGATCCTGAA................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................TTGTGATGATGTTGATCCTGAAa.................................................................................................................................. | 23 | a | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .AGTAGACTGCTCTGGGCAGAGT................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................TTGTGATGATGTTGATCCTGAACAGAC.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GAGTAGACTGCTCTGGGCAGAGTCACTCACATACAGTGCAAGCTTATTTCCCAGAAGGGTAAATACACCTTCAGTGGTTTCCTAAACACTGAACATTTTGTGATGATGTTGATCCTGAACAGACAGGTCTGTCGGGGTCATTTATCACGTGTTGGTTTGTTGTTTTTACAGTTCTTGAAGGTTTCTTTGTTTATTGTCAGGTGCACACTGCCTGCAAAGATGTGTACCATCCCGTCTGTCCCCTTGGCCA ....................................................................................................((((((...((((((((.(((((....))))))))))))).))))))....................(((((...(((((....)))))...)))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................CACATACAGTGCAAgacc................................................................................................................................................................................................................. | 18 | gacc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |