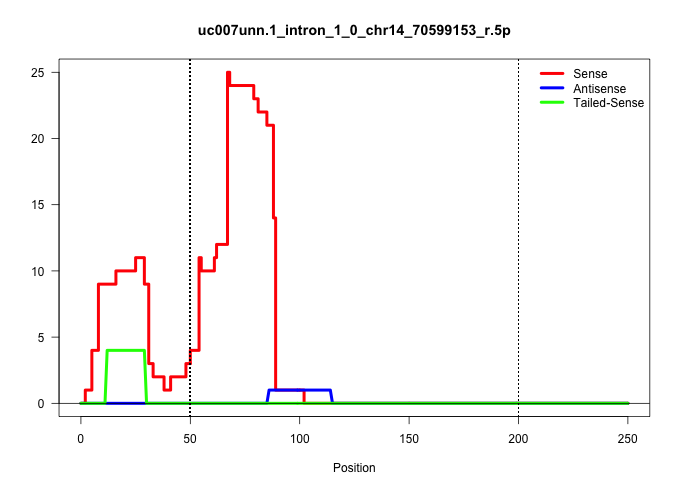
| Gene: Sorbs3 | ID: uc007unn.1_intron_1_0_chr14_70599153_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| TCAGCCACATGGACCAAGGACAGCAAGCGACAGGACAAGCGCTGGGTGAAGTACGAGGGAATCGGGCCCGTGGATGAGAGCGGCATGCCCATTGCCCCCCGATCTGTGAGTCCAGGGCTGAGGGTCCCAGACAGAGGGGGCCCAGAGACCTCCCAAGCTCTATCTCTAGGAGTATAATAGGGGTGGGGCACAGAGGCCGTACCCACACCTTGTGTGTGGAGTCACATAGATGTAATATGATACAGATTTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................CCGTGGATGAGAGCGGCATGCC................................................................................................................................................................. | 22 | 1 | 13.00 | 13.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GAGGGAATCGGGCCCGTGGATGAGAGCGGCATGC.................................................................................................................................................................. | 34 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATGGACCAAGGACAGCAAGCGAC........................................................................................................................................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 |
| ............ACCAAGGACAGCAAGCtg............................................................................................................................................................................................................................ | 18 | tg | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CACATGGACCAAGGACAGCAAGCGAC........................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| .....CACATGGACCAAGGACAGCAAGCG............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................CTGGGTGAAGTACGAGGGAATCGGGCC...................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................CGGGCCCGTGGATGAGAGC......................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AGCGACAGGACAAGCGCTGGGTGAAGTACG................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................TCGGGCCCGTGGATGAGAGCGGCA..................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTACGAGGGAATCGGGCCCGTGGATGAGAGCGGCATGCCCATTGCCCCCCGA.................................................................................................................................................... | 52 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................AAGTACGAGGGAATCGGGCCCGTGGATGAGA........................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........ATGGACCAAGGACAGCAAGCG............................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................AGGACAGCAAGCGACAGGACAA.................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..AGCCACATGGACCAAGGACAGCAAGCGACAG......................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TCAGCCACATGGACCAAGGACAGCAAGCGACAGGACAAGCGCTGGGTGAAGTACGAGGGAATCGGGCCCGTGGATGAGAGCGGCATGCCCATTGCCCCCCGATCTGTGAGTCCAGGGCTGAGGGTCCCAGACAGAGGGGGCCCAGAGACCTCCCAAGCTCTATCTCTAGGAGTATAATAGGGGTGGGGCACAGAGGCCGTACCCACACCTTGTGTGTGGAGTCACATAGATGTAATATGATACAGATTTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................GCCCATTGCCCCCCGATCTGTGAGTCCAG....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................AGCTCTATCTCTAGatt................................................................................. | 17 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |