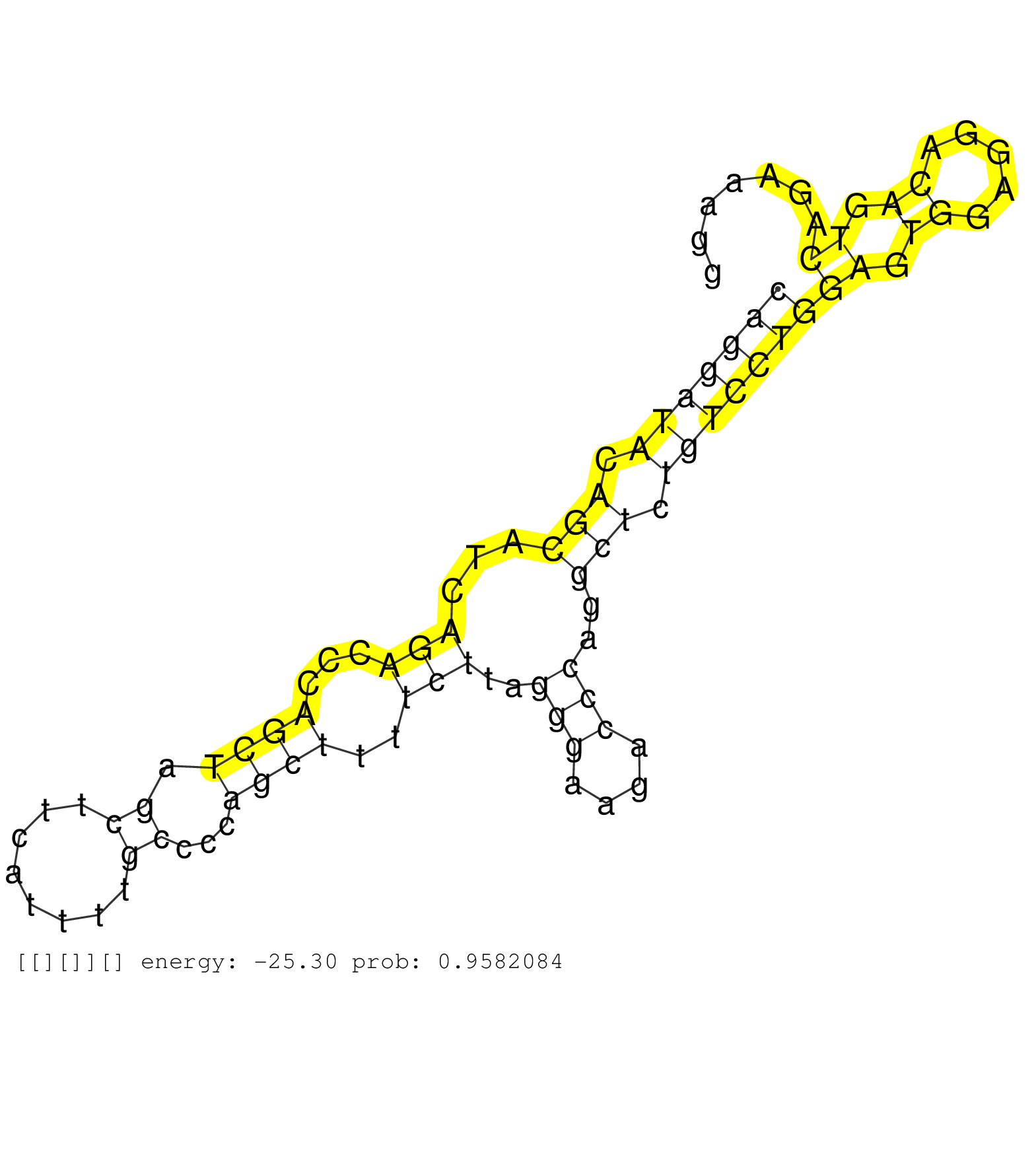
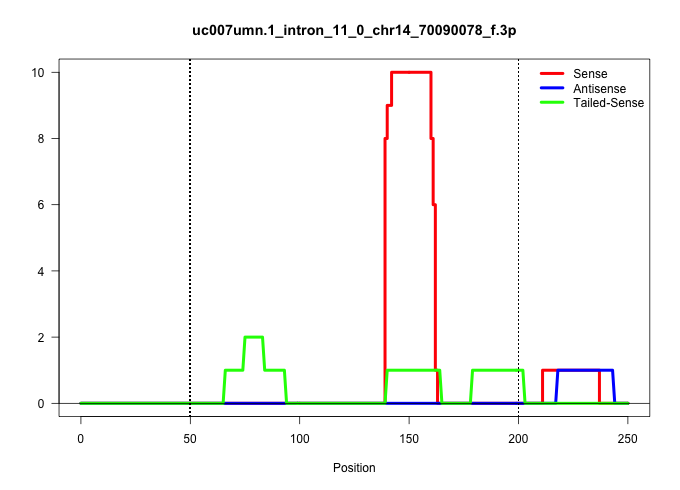
| Gene: Loxl2 | ID: uc007umn.1_intron_11_0_chr14_70090078_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| TGGCACATATCCTGGGCAAATTGTGGTTAGCAGGCCAGGCACAGGCTTTGCAGAGGGGCCCCTAACTGGCCAGGATACAGCATCAGACCCAGCTAGCTTCATTTTGCCCCAGCTTTTCTTAGGGAAGACCCAGGCTCTGTCCTGGAGTGGAGGACAGTCAGAAAGGCAGTTGTAGCATTCTCTGTCTTGATCCTCTCCAGGTTGTCATTAACCCCAACTATGAAGTGCCAGAATCAGATTTCTCTAACAA ......................................................................(((((((.(((...(((...((((.((........))...))))..)))..(((....)))..))).)))))))((.((.....)).))........................................................................................... ......................................................................71.............................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................TCCTGGAGTGGAGGACAGTCAGA........................................................................................ | 23 | 1 | 15.00 | 15.00 | 7.00 | - | 1.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ............................................................................................................................................CCTGGAGTGGAGGACAGTCAGA........................................................................................ | 22 | 1 | 12.00 | 12.00 | 7.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCCTGGAGTGGAGGACAGTCAG......................................................................................... | 22 | 1 | 10.00 | 10.00 | - | 8.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................TCCTGGAGTGGAGGACAGTCAGAA....................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCTGTCTTGATCCTCTCCAGttt............................................... | 24 | ttt | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCTGTCTTGATCCTCTCCAGatt............................................... | 24 | att | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCCTGGAGTGGAGGACAGTCA.......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTCTGTCTTGATCCTCTCCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTGGAGGACAGTCAGA........................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCAGGATACAGaggg...................................................................................................................................................................... | 18 | aggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTGGAGTGGAGGACAGTCAGAAct..................................................................................... | 25 | ct | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................CCCCAACTATGAAGTGCCAGAATCAG............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................TACAGCATCAGACCCAttt............................................................................................................................................................ | 19 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................TCCTGGAGTGGAGGACAGTCAGt........................................................................................ | 23 | t | 1.00 | 10.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTGGAGTGGAGGACAGTCAGAA....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGGCACATATCCTGGGCAAATTGTGGTTAGCAGGCCAGGCACAGGCTTTGCAGAGGGGCCCCTAACTGGCCAGGATACAGCATCAGACCCAGCTAGCTTCATTTTGCCCCAGCTTTTCTTAGGGAAGACCCAGGCTCTGTCCTGGAGTGGAGGACAGTCAGAAAGGCAGTTGTAGCATTCTCTGTCTTGATCCTCTCCAGGTTGTCATTAACCCCAACTATGAAGTGCCAGAATCAGATTTCTCTAACAA ......................................................................(((((((.(((...(((...((((.((........))...))))..)))..(((....)))..))).)))))))((.((.....)).))........................................................................................... ......................................................................71.............................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................CTTCATTTTGCCCCAGCTTTTCTTAt................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................................TATGAAGTGCCAGAATCAGATTTCTC...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AACTGGCCAGGATAgtca............................................................................................................................................................................. | 18 | gtca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |