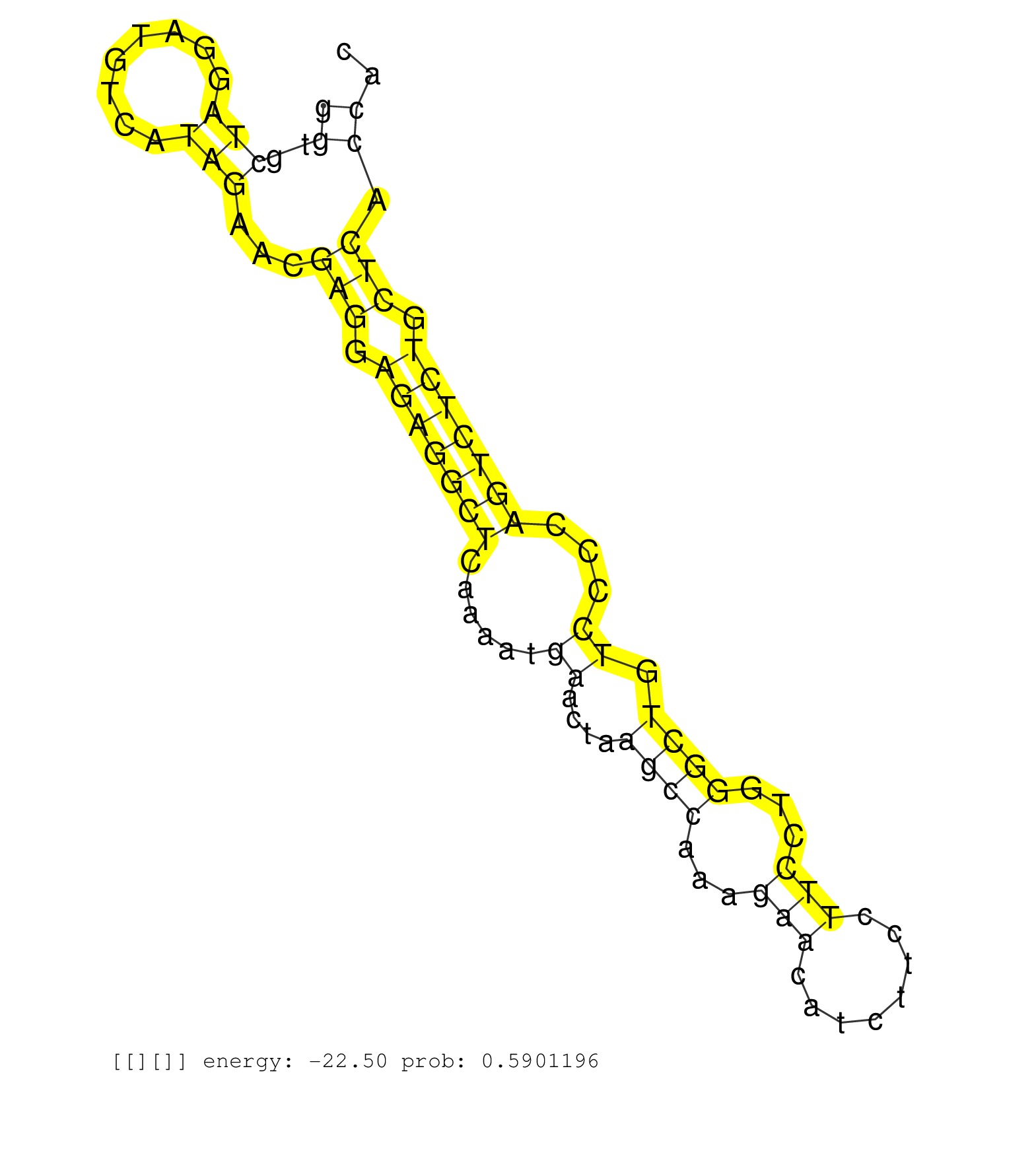

| Gene: Ints9 | ID: uc007uiw.1_intron_15_0_chr14_65656321_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TGCTGAGTGGCTCCATCCCCGTAGAGCAGTTTGTGCAGACCCTGGAGAAGGTGAGCTGGTGGCCTGCTCTGCCCCGAAGGGGGTGCTAGGATGTCATAGAACGAGGAGAGGCTCAAAATGAACTAAGCCAAAGAACATCTTCCTTCCTGGGCTGTCCCCAGTCTCTGCTCACCACCAGCCCTGTCTTCAGTCTCCAGCTCTGGAGCAGCAGGAATAGCACAGCACAGGCTGCCTGTCCTCCCTGGAGCCA .................................................................................((..(((........)))...(((.(((((((......((....((((...(((........)))...)))).))...))))))).))).))............................................................................. .................................................................................82...........................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TAGGATGTCATAGAACGAGGAGAGGCTC........................................................................................................................................ | 28 | 1 | 23.00 | 23.00 | 7.00 | 3.00 | 1.00 | 3.00 | - | 4.00 | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................TAGGATGTCATAGAACGAGGAGAGGCT......................................................................................................................................... | 27 | 1 | 21.00 | 21.00 | 8.00 | 8.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................TAGGATGTCATAGAACGAGGAGAGGC.......................................................................................................................................... | 26 | 1 | 14.00 | 14.00 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAAC............................................................................................................................... | 27 | 1 | 13.00 | 13.00 | 5.00 | 2.00 | - | - | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAACT.............................................................................................................................. | 28 | 1 | 9.00 | 9.00 | 2.00 | 2.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGAACTAAGCCAAAGAACATCTTCCTTC........................................................................................................ | 28 | 1 | 8.00 | 8.00 | 2.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTCATAGAACGAGGAGAGGCTCAAA..................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAACTA............................................................................................................................. | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TCCCCGTAGAGCAGTTTGTGCAGACCCT............................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................TGCTAGGATGTCATAGAACGAGGAGA............................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................AGGATGTCATAGAACGAGGAGAGGC.......................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTCATAGAACGAGGAGAGGCTCAAAA.................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCCATCCCCGTAGAGCAGTTTGTGCAG.................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAA................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GAGCAGTTTGTGCAGACCCTGGAGA.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................TGTCATAGAACGAGGAGAGGCTCAAAAT................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAGTGGCTCCATCCCCGTAGAGCAGT............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................GTAGAGCAGTTTGTGCAGACCCTGGA............................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TAGGATGTCATAGAACGAGGAGAGGCTa........................................................................................................................................ | 28 | a | 1.00 | 21.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................CCGTAGAGCAGTTTGTGCAGACC................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGGGCTGTCCCCAGTgcg..................................................................................... | 18 | gcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGA................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAACTAAGC.......................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................GTAGAGCAGTTTGTGCAGACCCTGG............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATGAAa............................................................................................................................... | 27 | a | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TAGGATGTCATAGAACGAGGAGAGGa.......................................................................................................................................... | 26 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................AGGATGTCATAGAACGAGGAGAGGCTC........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............TCCCCGTAGAGCAGTTTGTGCAGACCC................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................GTAGAGCAGTTTGTGCAGAtcct............................................................................................................................................................................................................... | 23 | tcct | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................AGGATGTCATAGAACGAGGAGAGGCTt........................................................................................................................................ | 27 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TCTGGAGCAGCAGGAATAGCACAGC........................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TAGAACGAGGAGAGGCTCAAAATG.................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GAACGAGGAGAGGCTCAAAATGAACTA............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................AGGATGTCATAGAACGAGGAGAGGCT......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGCAGACCCTGGAGAAGGTGAGCTGG............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTCATAGAACGAGGAGAGGCTCAAAt.................................................................................................................................... | 27 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GAGCAGTTTGTGCAGACCCTGGAGAA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCCTGGGCTGTCCCCAGTCTCTGCTCA............................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGAACTAAGCCAAAGAACATCTTCCTTCC....................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCTGAGTGGCTCCATCCCCGTAGAGCAGTTTGTGCAGACCCTGGAGAAGGTGAGCTGGTGGCCTGCTCTGCCCCGAAGGGGGTGCTAGGATGTCATAGAACGAGGAGAGGCTCAAAATGAACTAAGCCAAAGAACATCTTCCTTCCTGGGCTGTCCCCAGTCTCTGCTCACCACCAGCCCTGTCTTCAGTCTCCAGCTCTGGAGCAGCAGGAATAGCACAGCACAGGCTGCCTGTCCTCCCTGGAGCCA .................................................................................((..(((........)))...(((.(((((((......((....((((...(((........)))...)))).))...))))))).))).))............................................................................. .................................................................................82...........................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|