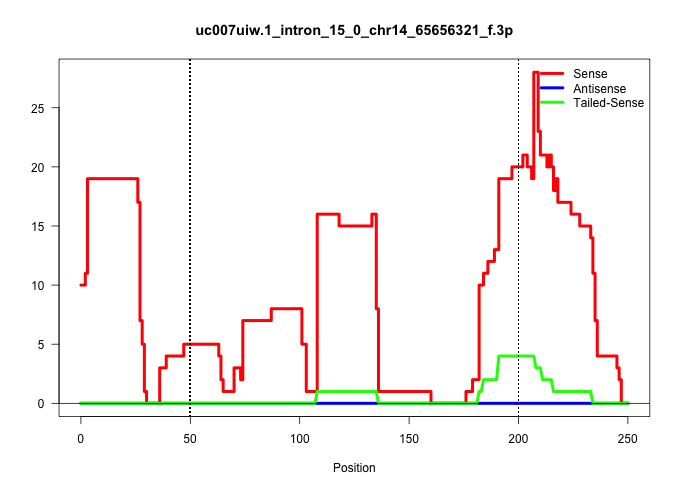
| Gene: Ints9 | ID: uc007uiw.1_intron_15_0_chr14_65656321_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TGATGGACTCAGGGCCTCAGAAAGGGTGGAGGAATCTGCTATTTTCCTGAGAGTTTTGTGCGTGGGGCCTTTCATAAATAAAACCTCTTGGTTGGACTCTGGCCCCGATGTCAGAGCTGGGCTGAGTGTAAACGCCATAGTGAGTGCTCAGTCAGGCCTTGCTCTCAGAGGCACTGTCCTCCTCTTTTCTCTTGGAACAGCATGGCTTCAGTGACATTAAGGTGGAAGACACAGCCAAGGGACATATTGT .............................................................................................................((((..((((.(((((((....((......))....)))))))((((((((.......))))).)))...(((...........))).)))).))))............................................ .......................................................................................................104.....................................................................................................208........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................TGTCAGAGCTGGGCTGAGTGTAAACGC................................................................................................................... | 27 | 1 | 8.00 | 8.00 | 1.00 | 1.00 | 3.00 | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................TGTCAGAGCTGGGCTGAGTGTAAACGCC.................................................................................................................. | 28 | 1 | 7.00 | 7.00 | 3.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTTTTCTCTTGGAACAGCATGGCTTC......................................... | 27 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAAATAAAACCTCTTGGTTGGACTCTGGC................................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGTGACATTAAGGTGGAAGACACAGC............... | 28 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...TGGACTCAGGGCCTCAGAAAGGGTGG............................................................................................................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGTGACATTAAGGTGGAAGACACAG................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGGACTCAGGGCCTCAGAAAGGG................................................................................................................................................................................................................................ | 23 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTCATAAATAAAACCTCTTGGTTGGACTCTG..................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTGGAACAGCATGGCTTCAGTGACAT................................. | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTGGAACAGCATGGCTTCAGTGACATT................................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGCTATTTTCCTGAGAGTTTTGTGCGTG.......................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTGGAACAGCATGGCTTCAGTGACA.................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGTGACATTAAGGTGGAAGACACAGCC.............. | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGGCTTCAGTGACATTAAGGTGGAAG...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................................TTAAGGTGGAAGACAgac................ | 18 | gac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................................TAAGGTGGAAGACACAGCCAAGGGACAT..... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAAATAAAACCTCTTGGTTGGACTCTG..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTGGAACAGCATGGCTTCAGTGAta.................................. | 25 | ta | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTTCTCTTGGAACAGCATGGCTTCA........................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAGTGACATTAAGGTGGAAGACACAGCC.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................TCTTGGAACAGCATGGCTTCAGTGAC................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TCCTCCTCTTTTCTCTTGGAACAGCATG.............................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................TGCTATTTTCCTGAGAGTTTTGTGCGT........................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGCATGGCTTCAGTGACATTAAGGTG.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...TGGACTCAGGGCCTCAGAAAGGGTGGA............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTTTTCTCTTGGAACAGCATGGCT........................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGTCAGAGCTGGGCTGAGTa.......................................................................................................................... | 20 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................................TTCTCTTGGAACAGCATGGCTTCAGTG..................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTTTTCTCTTGGAACAGCATGGCTTCA........................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGTCAGAGCTGGGCTGAGTGTAAACGCt.................................................................................................................. | 28 | t | 1.00 | 8.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGAGAGTTTTGTGCGTGGGGCCTTTC................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GCCATAGTGAGTGCTCAGTCAGGCCTT.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................TTGGTTGGACTCTGGCCCCGATGTCAGAGCT.................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTTCTCTTGGAACAGCATGGCTTCAt....................................... | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CATTAAGGTGGAAGACACAGCCAAGGGACATAT... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ATGGACTCAGGGCCTCAGAAAGGGTG.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCAGTGACATTAAGGTGGAAGACACA................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TAAGGTGGAAGACACAGCCAAGGGACATA.... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................TTGGAACAGCATGGCTTCAGTGACg.................................. | 25 | g | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TATTTTCCTGAGAGTTTTGTGCGTGG......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGGACTCAGGGCCTCAGAAAGGGT............................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCTCTTTTCTCTTGGAACAGCATGGC............................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTTTTCTCTTGGAACAGCATGGCct.......................................... | 26 | ct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTTTTCTCTTGGAACAGCATGGCTT.......................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TAAGGTGGAAGACACAGCCAAGGGACATAT... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TGATGGACTCAGGGCCTCAGAAAGGGTGGAGGAATCTGCTATTTTCCTGAGAGTTTTGTGCGTGGGGCCTTTCATAAATAAAACCTCTTGGTTGGACTCTGGCCCCGATGTCAGAGCTGGGCTGAGTGTAAACGCCATAGTGAGTGCTCAGTCAGGCCTTGCTCTCAGAGGCACTGTCCTCCTCTTTTCTCTTGGAACAGCATGGCTTCAGTGACATTAAGGTGGAAGACACAGCCAAGGGACATATTGT .............................................................................................................((((..((((.(((((((....((......))....)))))))((((((((.......))))).)))...(((...........))).)))).))))............................................ .......................................................................................................104.....................................................................................................208........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|