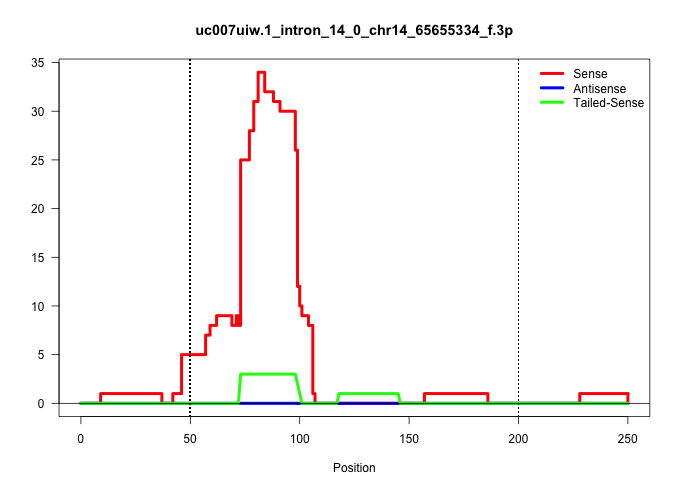
| Gene: Ints9 | ID: uc007uiw.1_intron_14_0_chr14_65655334_f.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(15) TESTES |
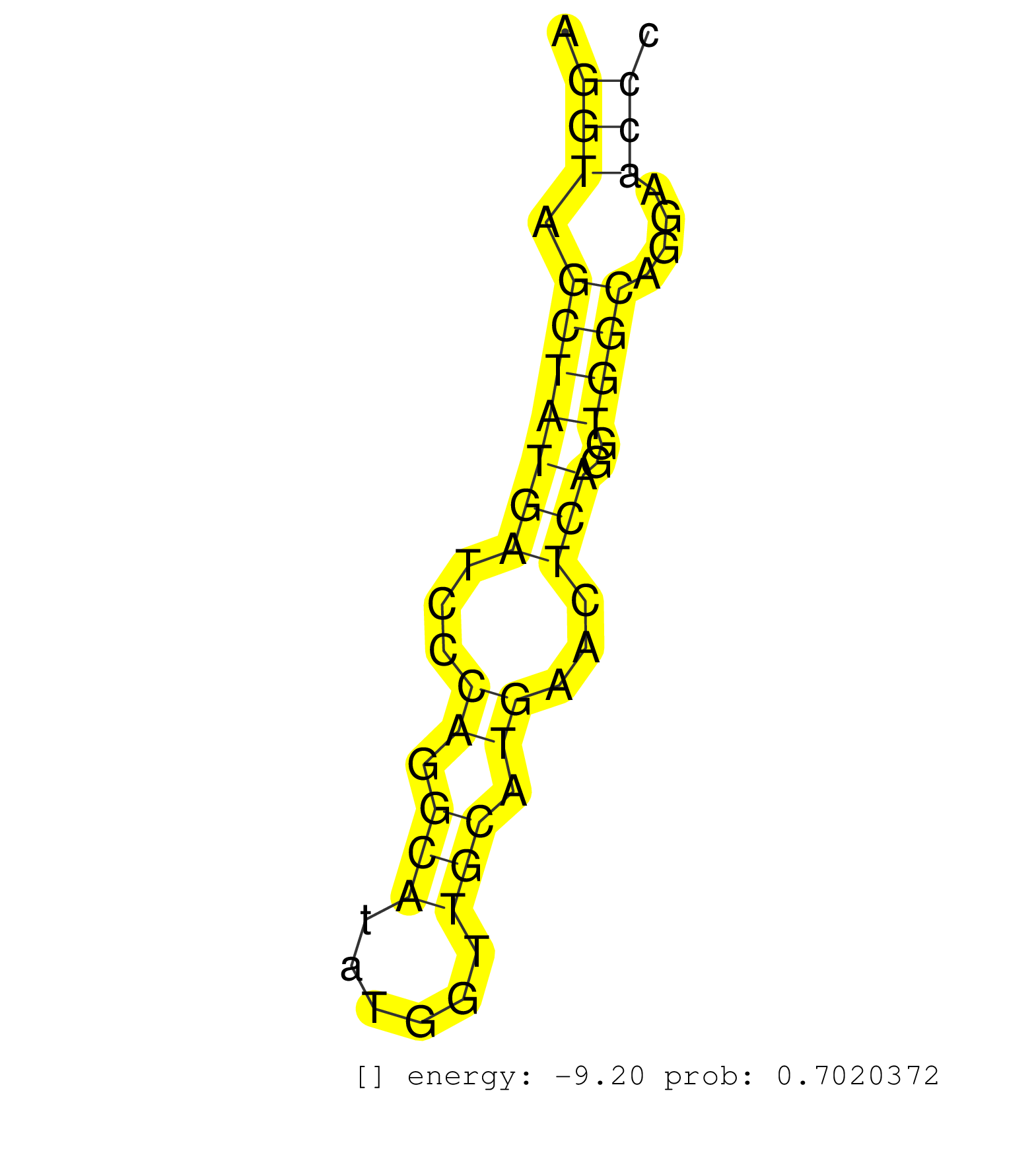
| GGATTTCATCAGCAAATGAATCCTGGAAATGCACACTGTTTCTTGCTGAAAGGTAGCTATGATCCCAGGCATATGGTTGCATGAACTCAGGTGGCAGGAACCCAGCTGCTGAGCCTGCTTTTCACGTCAGGCCAGCAGGTCACGGCCACGGAGTCAGTGAAGAGGCACTCACCAAGAGCCACACTTTCCTTCTGCCCTAGCCCCCTCCCAAGCCCACCCAGCCCACCAGCAGTAAGAAGAGGAAGCGGGT ...................................................(((.(((((((...((.(((......))).))...)))..))))....))).................................................................................................................................................... ..................................................51..................................................103................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGA....................................................................................................................................................... | 26 | 1 | 13.00 | 13.00 | 4.00 | - | - | 2.00 | 2.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | 1.00 |
| .......................................................................TATGGTTGCATGAACTCAGGTGGCAGG........................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - |
| ...............................................................................CATGAACTCAGGTGGCAGGAACCCAGC................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................TATGATCCCAGGCATATGGTTGCATGA...................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGAA...................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TGAAAGGTAGCTATGATCCCAGGCA................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGGAAATGCACACTGTagg................................................................................................................................................................................................................ | 19 | agg | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................TGAACTCAGGTGGCAGGAACCCAGC................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGG........................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGCATGAACTCAGGTGGCAGGAACCCAGC................................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGt....................................................................................................................................................... | 26 | t | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TGAAAGGTAGCTATGATCCCAGGCAT.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................GCAGTAAGAAGAGGAAGCGGGT | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TCCCAGGCATATGGTTGCATGAACTCAGG............................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CAGCAAATGAATCCTGGAAATGCACACT..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGAAC..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................TTTTCACGTCAGGCCAGCAGGTCAaggc........................................................................................................ | 28 | aggc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................TTGCTGAAAGGTAGCTATGATCCCAGG..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TATGGTTGCATGAACTCAGGTGGCAGGA....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGCATGAACTCAGGTGGCAGGAACCCA.................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGAAGAGGCACTCACCAAGAGCCACACTT................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................TGATCCCAGGCATATGGTTGCATGAACTC.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGAt...................................................................................................................................................... | 27 | t | 1.00 | 13.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TGAAAGGTAGCTATGATCCCAGGCATA................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGAACTCAGGTGGCAGGAACCCAGCT............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGGTTGCATGAACTCAGGTGGCAGGAAt..................................................................................................................................................... | 28 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGATTTCATCAGCAAATGAATCCTGGAAATGCACACTGTTTCTTGCTGAAAGGTAGCTATGATCCCAGGCATATGGTTGCATGAACTCAGGTGGCAGGAACCCAGCTGCTGAGCCTGCTTTTCACGTCAGGCCAGCAGGTCACGGCCACGGAGTCAGTGAAGAGGCACTCACCAAGAGCCACACTTTCCTTCTGCCCTAGCCCCCTCCCAAGCCCACCCAGCCCACCAGCAGTAAGAAGAGGAAGCGGGT ...................................................(((.(((((((...((.(((......))).))...)))..))))....))).................................................................................................................................................... ..................................................51..................................................103................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................CCACCAGCAGTAAGAagc............. | 18 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TCACCAAGAGCCACAaccg................................................................... | 19 | accg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |