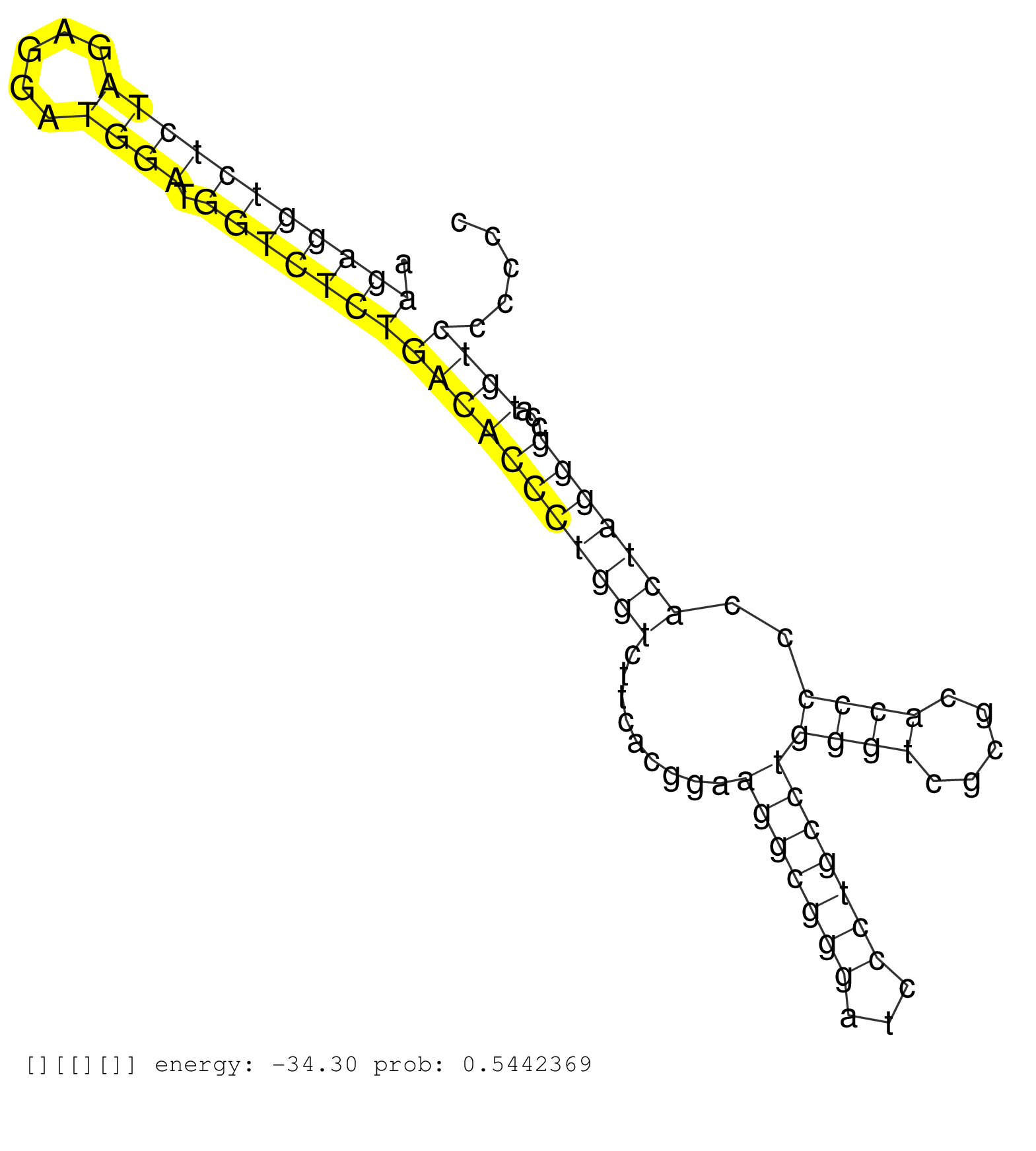

| Gene: Fdft1 | ID: uc007uhi.1_intron_6_0_chr14_63783467_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| TGCGATTCCGCATGGGAGGCCGGCGGAATTTTATACCCAAGATGGACCAGGTAAGTCGAGCATCCCTGCTCCCTCGCAGGCCTGTGGCGGCTTCCCGCACAAGAGGTCTCTAGAGGATGGATGGTCTCTGACACCCTGGTCTTCACGGAAGGCGGGATCCCTGCCTGGGTCGCGCACCCCCACTAGGGCCATGTCCCCCCCCCCCCCCCCCCGCCTTTCAAGGTTTCTGTCTGTTCAAGCCATTGCTTTT .....................................................................................................(((((((((((.....)))).)))))))(((((((((((.........(((((((...)))))))((((.....))))..)))))))...))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCC.................................................................................................................. | 26 | 1 | 6.00 | 6.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCT................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCTG................................................................................................................ | 28 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCTAGAGGATGGATGGTCTCTGACACCC.................................................................................................................. | 28 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACAC.................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACC................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................TGGTCTCTGACACCCagcc.............................................................................................................. | 19 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................CCCAAGATGGACCAGGTAAGTCGAGC............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACcccc.................................................................................................................. | 26 | cccc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTATACCCAAGATGGACCAGG....................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCTAGAGGATGGATGGTCTCTGACACC................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGGATGGTCTCTGACACCCTGGTCTTC.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCTGGTC............................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGCTCCCTCGCAGGCCTGTGGCGGCTTC............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCac................................................................................................................ | 28 | ac | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GGCGGAATTTTATACCCAAGATGGACCA......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTGACACCCTGGTCTTCACGGAAGGC................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCTGa............................................................................................................... | 29 | a | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GCGGAATTTTATACCCAAGATGGACCAGG....................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CGGCGGAATTTTATACCCAAGATGGACC.......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAGAGGATGGATGGTCTCTGACACCCTGG............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................TGCTCCCTCGCAGGCCTGTGGCGGCTTtc........................................................................................................................................................... | 29 | tc | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGCTCCCTCGCAGGCCTGTGGCGGCTT............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GCGGAATTTTATACCCAAGATGGACCAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........ATGGGAGGCCGGCGGAATTTTATACCC.................................................................................................................................................................................................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ............TGGGAGGCCGGCGGAATTT........................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GGGAGGCCGGCGGAATTTTAT........................................................................................................................................................................................................................ | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| TGCGATTCCGCATGGGAGGCCGGCGGAATTTTATACCCAAGATGGACCAGGTAAGTCGAGCATCCCTGCTCCCTCGCAGGCCTGTGGCGGCTTCCCGCACAAGAGGTCTCTAGAGGATGGATGGTCTCTGACACCCTGGTCTTCACGGAAGGCGGGATCCCTGCCTGGGTCGCGCACCCCCACTAGGGCCATGTCCCCCCCCCCCCCCCCCCGCCTTTCAAGGTTTCTGTCTGTTCAAGCCATTGCTTTT .....................................................................................................(((((((((((.....)))).)))))))(((((((((((.........(((((((...)))))))((((.....))))..)))))))...))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................TCTGTTCAAGCCATTGCaca.... | 20 | aca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................TCTCTAGAGGATGGATGG.............................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ........................................................................................................TCTCTAGAGGATGGATGGtt.............................................................................................................................. | 20 | tt | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |