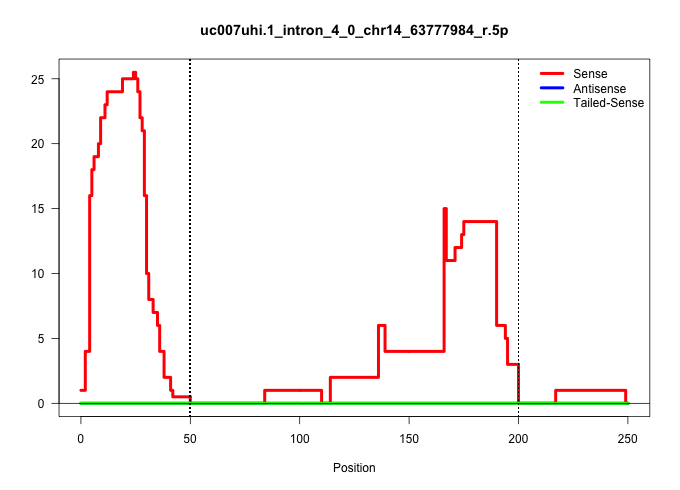
| Gene: Fdft1 | ID: uc007uhi.1_intron_4_0_chr14_63777984_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(21) TESTES |
| TCACTGAGAGCAAGGAGAAGGACCGACAAGTGCTGGAGGACTTCCCCACGGTACTGGGCTCTCTGGGCATAGGCTGTCGATAATTGCCAGTGCAGTTGTCGGTACTGGGCTCTGGGCATAGGCTGTTGATAATTGGCAGTGCAGTTGTCGGCCCCTGAGCTGAGAGAAAGGTGATGTGAACACTCTGAGGGCAGCTGAGAGTCCAAAGCCAGACTTTTTCTGCTTGGAATCCTAACTTGGTAATTGGTGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................AAAGGTGATGTGAACACTCTGAGG............................................................ | 24 | 1 | 8.00 | 8.00 | - | 3.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ....TGAGAGCAAGGAGAAGGACCGACAAG............................................................................................................................................................................................................................ | 26 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CAGTGCAGTTGTCGGCCCCTGAGCTGAGAGA................................................................................... | 31 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGAGAGCAAGGAGAAGGACCGACAA............................................................................................................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................AAAGGTGATGTGAACACTCTGAGGGCAGCTGAGA.................................................. | 34 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GGCATAGGCTGTTGATAATTGGCAG............................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ACTGAGAGCAAGGAGAAGGACCGAC............................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ...........AAGGAGAAGGACCGACAAGTGCTGGAG.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........GCAAGGAGAAGGACCGACAAGTGCTGG...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GCAAGGAGAAGGACCGACAAGTGCTGGAGGAC................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............AGGAGAAGGACCGACAAGTGCTGG...................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTCTGCTTGGAATCCTAACTTGGTAATTGGTG. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGTGAACACTCTGAGGGCAGC....................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGAGAGCAAGGAGAAGGACCGACAAGT........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........AGCAAGGAGAAGGACCGACAAGTGCTG....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AGAGCAAGGAGAAGGACCGACAAGTGC......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GAGAGCAAGGAGAAGGACCGACAA............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ACTGAGAGCAAGGAGAAGGACCGACAAGT........................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGTGAACACTCTGAGGGCAG........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCAGTGCAGTTGTCGGTACTGGGC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................GGACCGACAAGTGCTGGAG.................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGAGAGCAAGGAGAAGGACCG................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTGAACACTCTGAGGGCAGC....................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....GAGAGCAAGGAGAAGGACCGACA.............................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................ACAAGTGCTGGAGGACT................................................................................................................................................................................................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ........................GACAAGTGCTGGAGGACTTCCCCACG........................................................................................................................................................................................................ | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCACTGAGAGCAAGGAGAAGGACCGACAAGTGCTGGAGGACTTCCCCACGGTACTGGGCTCTCTGGGCATAGGCTGTCGATAATTGCCAGTGCAGTTGTCGGTACTGGGCTCTGGGCATAGGCTGTTGATAATTGGCAGTGCAGTTGTCGGCCCCTGAGCTGAGAGAAAGGTGATGTGAACACTCTGAGGGCAGCTGAGAGTCCAAAGCCAGACTTTTTCTGCTTGGAATCCTAACTTGGTAATTGGTGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|