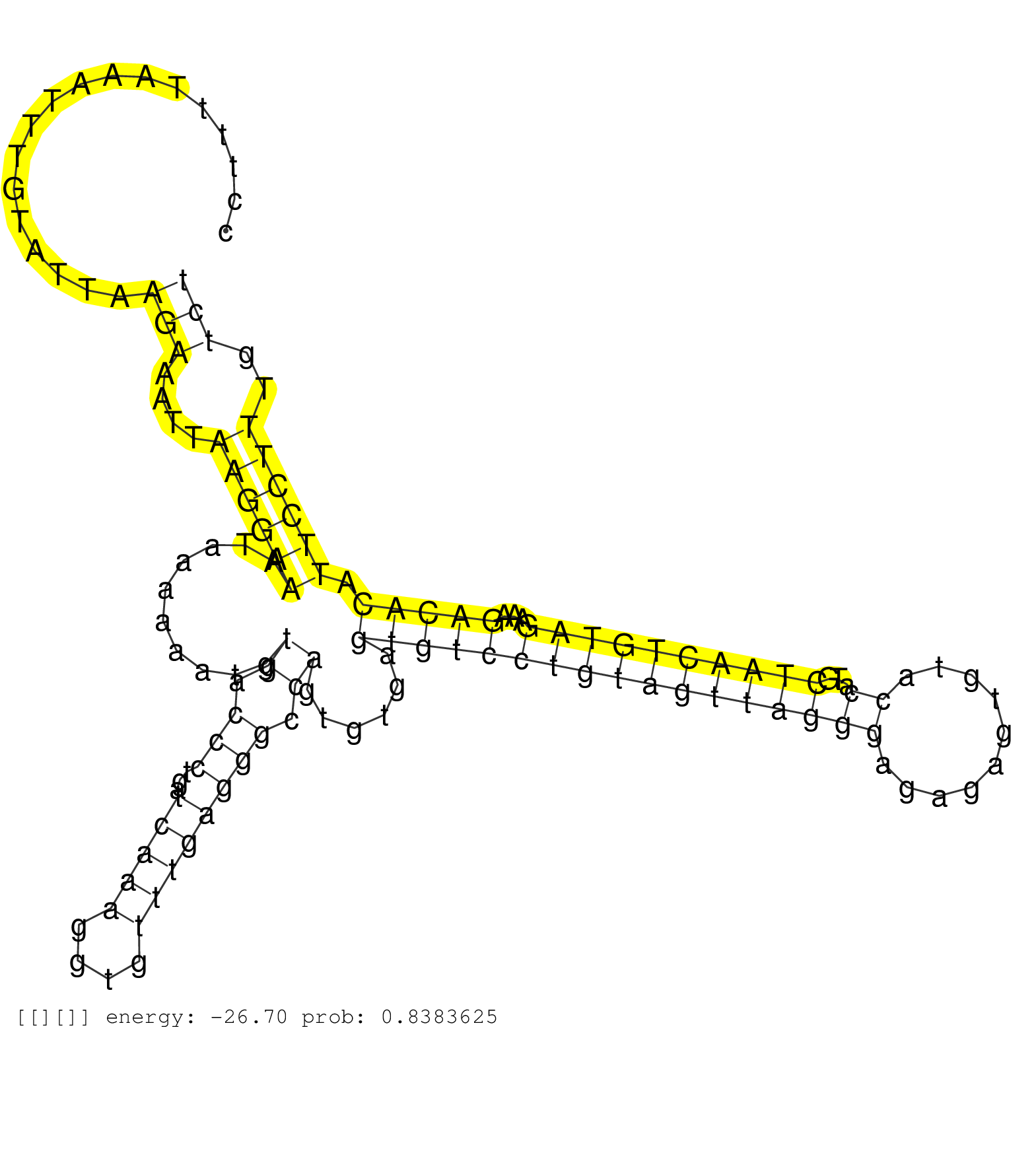

| Gene: Cdadc1 | ID: uc007uep.1_intron_0_0_chr14_60186876_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| TGGATGTCTTATCATTTACAGTCTGGAAAAATACATAATAACTACATTTTCCTTTTAAATTTGTATTAAGAAATTAAGGAAATAAAAAATGTGACCCTGATCAAAGGTGTTTGAGGGCCAGTGTGAGTGTCCTGTAGTTAGGGAGAGAGTGTACCATGCTAACTGTAGAAAGACACATTCCTTTGTCTGTTGAAATTAAGAGCATCTGTCCATCAAAAGATCCCACTAAGAGGGTGTAAAGGCAAGCCAC ....................................................................(((....((((((..........((.(((...(((((....)))))))).))......(((((((((((((((((..........))...))))))))))...))))).))))))..))).............................................................. ..................................................51.......................................................................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCTTT.................................................................. | 28 | 1 | 13.00 | 13.00 | 4.00 | 3.00 | 4.00 | - | - | - | 1.00 | 1.00 | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCTT................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TAACTGTAGAAAGACACATTCCTTTGTC............................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TAAGAGGGTGTAAAGGCAAGCCACggat | 28 | ggat | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCTTTGT................................................................ | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................GGGAGAGAGTGTACCATGCTAACTGTAGAAAG.............................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCT.................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCTTTGTC............................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................CCATGCTAACTGTAGAAAGACACATTCCT.................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTGTAGTTAGGGAGAGAGTGTACC............................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATTCCTTTGTCTGTTGAAATT..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCTTTG................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TAACTGTAGAAAGACACATTCCTTTGT................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACCATGCTAACTGTAGAAAGACACATTC...................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GCTAACTGTAGAAAGACACATTCCTTTGT................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGTCTGTTGAAATTAAGAGCATCTGTCCA...................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................TAAGAGCATCTGTCCATCAAAAGATCCC.......................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................TAAATTTGTATTAAGAAATTAAGGAAAT....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................TAAGAGGGTGTAAAGGCAAGCC.. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................TACCATGCTAACTGTAGAAAGACACATTCCT.................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGTCTGTTGAAATTAAGAGCATCTGTCCATC.................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGCTAACTGTAGAAAGACACATTCCattt................................................................. | 29 | attt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CATTCCTTTGTCTGTTGAAATTAAGAGCATC............................................ | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............TTACAGTCTGGAAAAATACATAATAACTAC............................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CCATGCTAACTGTAGAAAGACACATTCCTTT.................................................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GTAGAAAGACACATTCCTTTGTCTGTTG.......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................GCTAACTGTAGAAAGACACATTCCTTTG................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGAAATTAAGAGCATCTGTCCATCAAA................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TAAGAGCATCTGTCCATCAAAAGATCCCAC........................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGGATGTCTTATCATTTACAGTCTGGAAAAATACATAATAACTACATTTTCCTTTTAAATTTGTATTAAGAAATTAAGGAAATAAAAAATGTGACCCTGATCAAAGGTGTTTGAGGGCCAGTGTGAGTGTCCTGTAGTTAGGGAGAGAGTGTACCATGCTAACTGTAGAAAGACACATTCCTTTGTCTGTTGAAATTAAGAGCATCTGTCCATCAAAAGATCCCACTAAGAGGGTGTAAAGGCAAGCCAC ....................................................................(((....((((((..........((.(((...(((((....)))))))).))......(((((((((((((((((..........))...))))))))))...))))).))))))..))).............................................................. ..................................................51.......................................................................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................GCATCTGTCCATCaga.................................... | 16 | aga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |