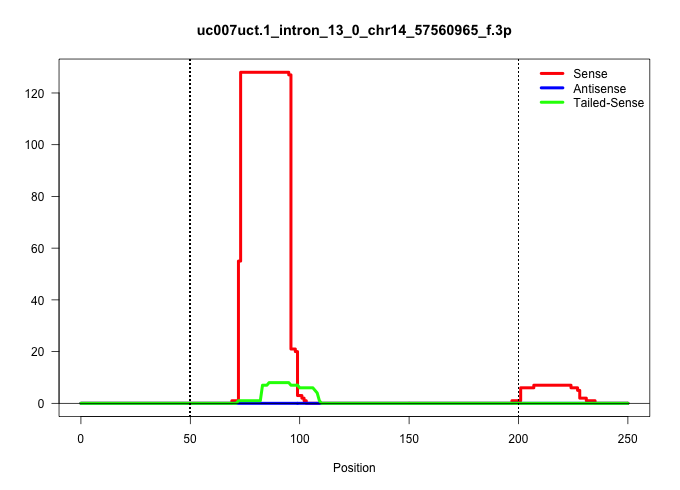
| Gene: Zmym2 | ID: uc007uct.1_intron_13_0_chr14_57560965_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(23) TESTES |
| CCCCCCTCCCCCCCCCCAAAAAAAAAAAACAAAAAAACCAAAACAAACCTATTATGGTGGCTCATACCTGTAAGCTCATGATGTAGGAGACTGAGGCAGGAGGATTGCCACAAGGTTAAGGCCAGCCTTTTTGACATGTTTAATGATTTTTTTTTAAAACAAAACCACATGTTGTTCTTTCCCTATGTCTTGTTTTTCAGATGAGAATTGGAAGACAGAGTATGTCCCAGTGCCTATTCCTGTCCCTGTG .....................................................................................................(((..((.((((((..(((..((((............(((((.((....)).)))))............))))..)))..))).)))...))..))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................GCTCATGATGTAGGAGACTGAGG.......................................................................................................................................................... | 23 | 1 | 65.00 | 65.00 | 20.00 | 9.00 | 19.00 | 9.00 | 2.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 |
| ........................................................................AGCTCATGATGTAGGAGACTGAGG.......................................................................................................................................................... | 24 | 1 | 42.00 | 42.00 | 14.00 | 13.00 | 3.00 | 2.00 | 2.00 | 1.00 | 1.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................AGCTCATGATGTAGGAGACTGAGGCAG....................................................................................................................................................... | 27 | 1 | 10.00 | 10.00 | 4.00 | 2.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GCTCATGATGTAGGAGACTGAGGCAG....................................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................AAGCTCATGATGTAGGAGACTGAGGCAG....................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAATTGGAAGACAGAGTATGTCCC...................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGAGACTGAGGCAGGAGGATccca............................................................................................................................................. | 26 | ccca | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGAGAATTGGAAGACAGA............................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGAGACTGAGGCAGGAGGATccat............................................................................................................................................. | 26 | ccat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGCTCATGATGTAGGAGACTGAGGCA........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAGACTGAGGCAGGAGGATTGata............................................................................................................................................ | 24 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................CAGATGAGAATTGGAAGACAGAGTATG.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGAGACTGAGGCAGGAGGATga............................................................................................................................................... | 24 | ga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGAGACTGAGGCAGGAGGATcca.............................................................................................................................................. | 25 | cca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGCTCATGATGTAGGAGACTGAGGCAGGAG.................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TTGGAAGACAGAGTATGTCCCAGTGCCT............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................GTAAGCTCATGATGTAGGAGACTGAGGCAGGA..................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TAGGAGACTGAGGaacg...................................................................................................................................................... | 17 | aacg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGCTCATGATGTAGGAGACTGAGa.......................................................................................................................................................... | 24 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AGCTCATGATGTAGGAGACTGAGGCAGGAGG................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAATTGGAAGACAGAGTATGTCCCAGT................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................GCTCATGATGTAGGAGACTGAG........................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAATTGGAAGACAGAGTATGTCC....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| CCCCCCTCCCCCCCCCCAAAAAAAAAAAACAAAAAAACCAAAACAAACCTATTATGGTGGCTCATACCTGTAAGCTCATGATGTAGGAGACTGAGGCAGGAGGATTGCCACAAGGTTAAGGCCAGCCTTTTTGACATGTTTAATGATTTTTTTTTAAAACAAAACCACATGTTGTTCTTTCCCTATGTCTTGTTTTTCAGATGAGAATTGGAAGACAGAGTATGTCCCAGTGCCTATTCCTGTCCCTGTG .....................................................................................................(((..((.((((((..(((..((((............(((((.((....)).)))))............))))..)))..))).)))...))..))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|