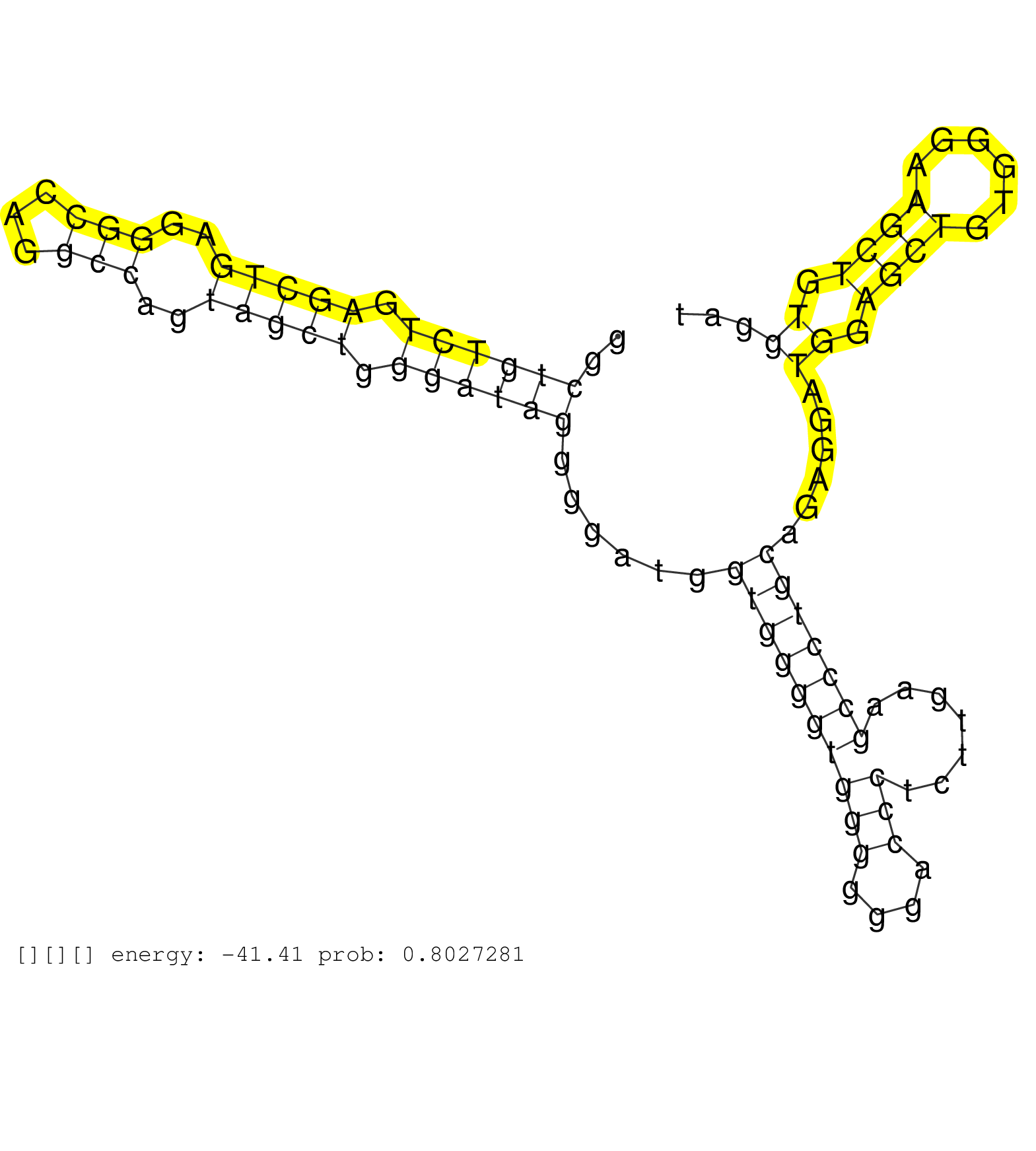
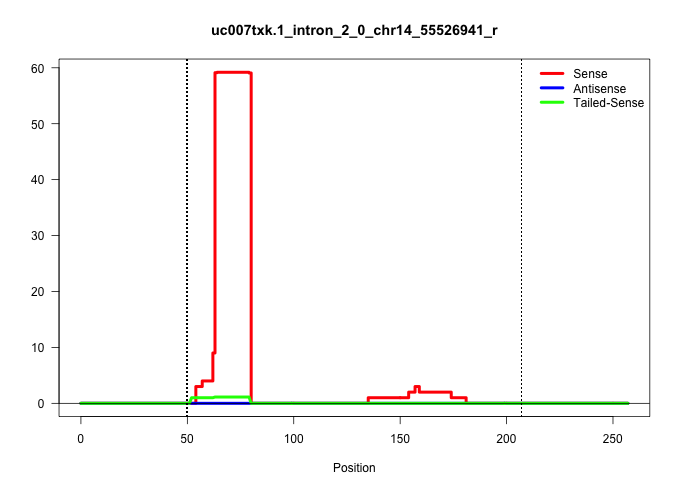
| Gene: mboct | ID: uc007txk.1_intron_2_0_chr14_55526941_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(14) TESTES |
| CTCAACTACCGCAACATCTGGAAAAATCTGCTTATCCTGGGCTTCACCAAGTGAGCCTGGCTGTCTGAGCTGAGGGCCAGGCCAGTAGCTGGGATAGGGGATGGTGGGGTGGGGGGACCCTCTTGAAGCCCTGCAGAGGATGGAGCTGTGGGAAGCTGTGGATTGGGGTTCAGACGCTGTTCTTTGCTTCCCCCACCTGCTGTGCAGCTTTATTGCCCATGCCATTCGCCACTGCTACCAGCCTGTGGGAGGAGGAG ............................................................((((((.(((((..(((...)))..))))).))))))......((((((((((....))).......)))))))......((.((((......)))).))................................................................................................. ..........................................................59......................................................................................................163............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................TCTGAGCTGAGGGCCAG................................................................................................................................................................................. | 17 | 2 | 71.00 | 71.00 | 16.00 | 22.00 | 18.00 | 4.50 | 2.00 | 3.50 | 3.00 | - | 1.50 | - | - | - | - | - | 0.50 |
| ..............................................................GTCTGAGCTGAGGGCCAG................................................................................................................................................................................. | 18 | 1 | 18.00 | 18.00 | 9.00 | 2.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................GCCTGGCTGTCTGAGCTGAGGGCCAG................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .............................................................................................................................................GGAGCTGTGGGAAGCTGTGGA............................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................TGGCTGTCTGAGCTGAGGGCCAG................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................GAGGATGGAGCTGTGGGAAGCTGT.................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GTGGATTGGGGTTCAGA................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GAGGATGGAGCTGTGGGAAGCTGTGGAT.............................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGTGGGAAGCTGTGGATTGGGGTT....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................GAGCCTGGCTGTCTGAGCTGAGGGCCcg................................................................................................................................................................................. | 28 | cg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................ACCAAGTGAGCCTGGCTGTCTGAGCTGAGGG..................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCTGTGGATTGGGGTTCAGACGCTGTT............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................TGTGGATTGGGGTTCAGACGCT............................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGGGAAGCTGTGGATT............................................................................................. | 16 | 6 | 0.17 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTGAGCTGAGGGCCAc................................................................................................................................................................................. | 17 | c | 0.12 | 0.12 | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - |
| ...............................................................TCTGAGCTGAGGGCCA.................................................................................................................................................................................. | 16 | 8 | 0.12 | 0.12 | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CTGAGCTGAGGGCCAG................................................................................................................................................................................. | 16 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - |
| CTCAACTACCGCAACATCTGGAAAAATCTGCTTATCCTGGGCTTCACCAAGTGAGCCTGGCTGTCTGAGCTGAGGGCCAGGCCAGTAGCTGGGATAGGGGATGGTGGGGTGGGGGGACCCTCTTGAAGCCCTGCAGAGGATGGAGCTGTGGGAAGCTGTGGATTGGGGTTCAGACGCTGTTCTTTGCTTCCCCCACCTGCTGTGCAGCTTTATTGCCCATGCCATTCGCCACTGCTACCAGCCTGTGGGAGGAGGAG ............................................................((((((.(((((..(((...)))..))))).))))))......((((((((((....))).......)))))))......((.((((......)))).))................................................................................................. ..........................................................59......................................................................................................163............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|