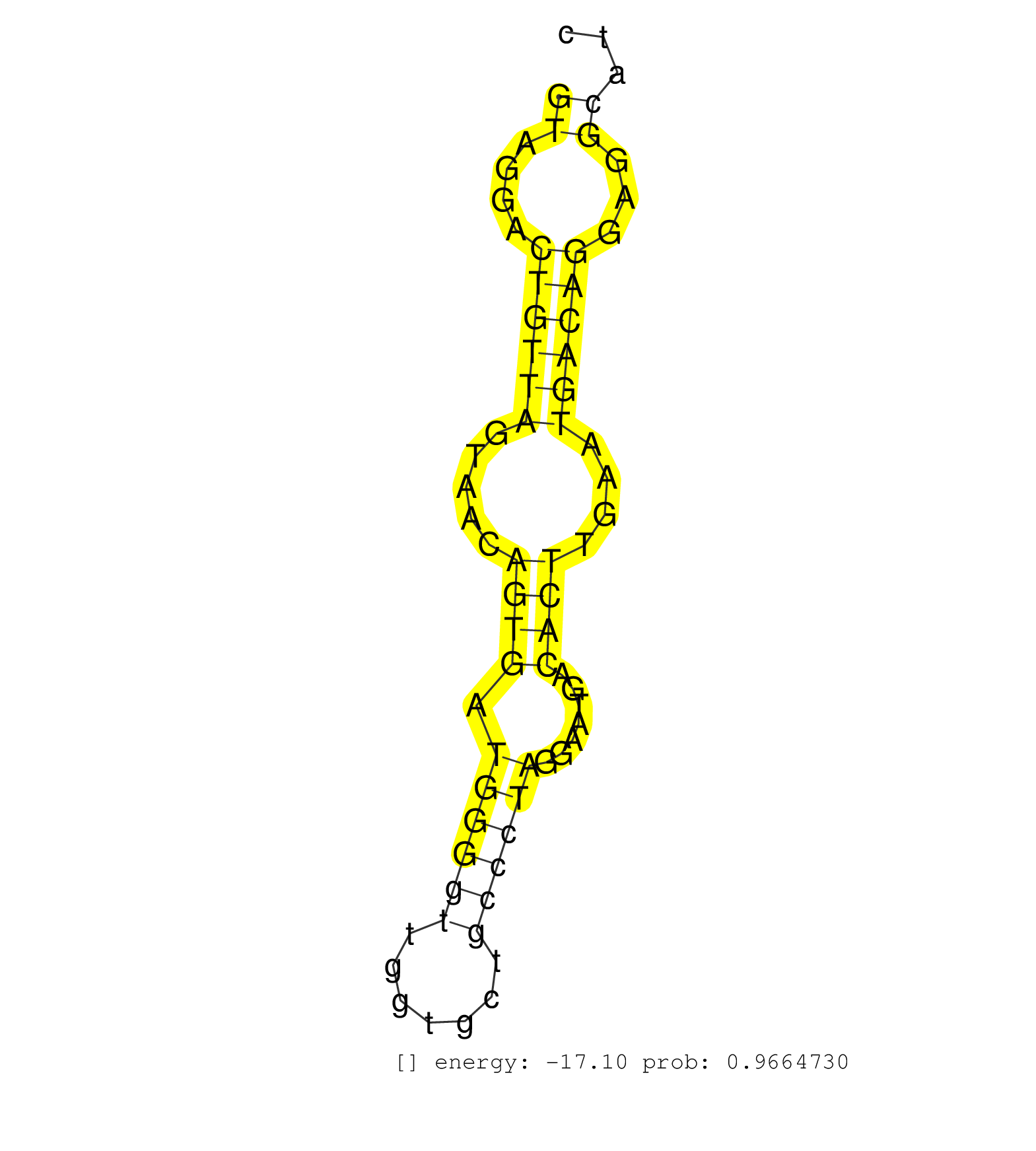

| Gene: Dlg7 | ID: uc007tid.1_intron_13_0_chr14_48031243_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| TGACCAAAGACAAACTTCTGAAAAACAACCATTAGACAGAGAGAGAAAAGGTAGGACTGTTAGTAACAGTGATGGGGTTGGTGCTGCCCTAGGAATGACACTTGAATGACAGGAGGCATCTGATGAGATGATTTTCCCCAGTTATGCAGCCTGTGCTGTTCACGTCAGGGAAAGGGACTGAATCAGCGGCT ..................................................((....((((((.....((((.((((((.......)))))).......))))....))))))...)).......................................................................... ..................................................51...................................................................120..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGACTGTTAGTAACAGTGATGGG................................................................................................................... | 26 | 1 | 33.00 | 33.00 | 6.00 | 2.00 | 5.00 | 1.00 | 3.00 | - | 1.00 | 2.00 | 3.00 | 1.00 | 2.00 | 2.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | 1.00 |
| ..................................................GTAGGACTGTTAGTAACAGTGATGGGG.................................................................................................................. | 27 | 1 | 10.00 | 10.00 | 3.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGTTAGTAACAGTGATGGGGTT................................................................................................................ | 29 | 1 | 7.00 | 7.00 | - | - | 1.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAGGACTGTTAGTAACAGTGATGGGGT................................................................................................................. | 28 | 1 | 5.00 | 5.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TAGGAATGACACTTGAATGACAGGAGGC.......................................................................... | 28 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGTTAGTAACAGTGATGG.................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GAAAGGGACTGAATCAGCGG.. | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TAGGAATGACACTTGAATGACAGGAGG........................................................................... | 27 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGACTGTTAGTAACAGTGATGGGGTT................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CACGTCAGGGAAAGGGACTGAATCAGCGG.. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACACTTGAATGACAGGAGGCATCTGA.................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGTTAGTAACAGTGATGGGt.................................................................................................................. | 27 | t | 2.00 | 33.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACACTTGAATGACAGGAGGCATC....................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAAAAACAACCATTAGACAGAGAGAa.................................................................................................................................................. | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................TAGGAATGACACTTGAATGACAGGAGGCA......................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CTGAATCAGCGGagga | 16 | agga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AGGGAAAGGGACTGAATCAGCGGCT | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TAGGAATGACACTTGAATGACAGGAG............................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAAAAACAACCATTAGACAGAGAGAGA................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TCACGTCAGGGAAAGGGACT............ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTAGGACTGTTAGTAACAGTGATGGGtt................................................................................................................. | 28 | tt | 1.00 | 33.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGACTGTTAGTAACAGTGATGGGGTTG............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................AGGACTGTTAGTAACAGTGATGGGGTT................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAAAAACAACCATTAGACAGAGAGAGt................................................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TAGGAATGACACTTGAATGACAGGAGGCt......................................................................... | 29 | t | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CTGATGAGATGATTgtac...................................................... | 18 | gtac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TGACCAAAGACAAACTTCTGAAAAACAACCATTAGACAGAGAGAGAAAAGGTAGGACTGTTAGTAACAGTGATGGGGTTGGTGCTGCCCTAGGAATGACACTTGAATGACAGGAGGCATCTGATGAGATGATTTTCCCCAGTTATGCAGCCTGTGCTGTTCACGTCAGGGAAAGGGACTGAATCAGCGGCT ..................................................((....((((((.....((((.((((((.......)))))).......))))....))))))...)).......................................................................... ..................................................51...................................................................120..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|