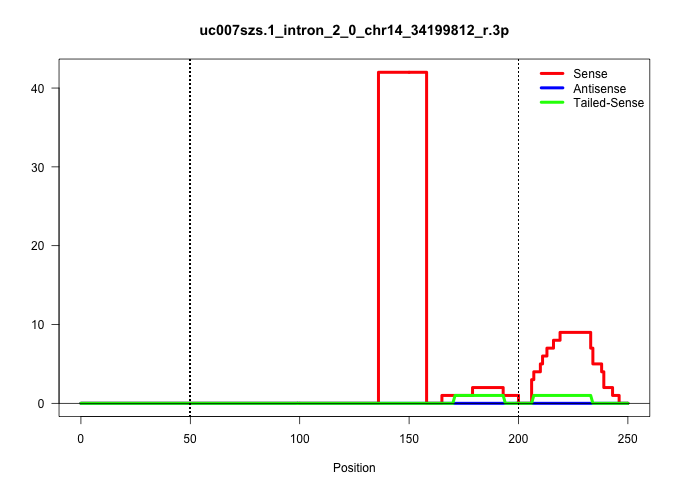
| Gene: Mapk8 | ID: uc007szs.1_intron_2_0_chr14_34199812_r.3p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(10) TESTES |
| AGAAAAAAGACAAACAGCTTATTACTTAGAAAGGAATCTTACAGAACTACTCTCCAGAAAGTTAAAGTTAATGATAACTGATTGACCTTTGTCTCATGATTCATTTTTTGCAAGTTGTATGTCTAGCTGATATTTCAAATAATTAATTGTATATATACATCCAGTAAACTATAACAGTTGTTCTGACTTGTGTCTTTCAGCCACCACCAAAGATCCCGGACAAGCAGTTAGATGAGAGGGAGCACACAAT ..........................................................................................................................................................(((((..(((..((((.....))))...)))...)))))......................................................... ...................................................................................................................................132.................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................AAATAATTAATTGTATATATACATCCAGTAAACTATAACAGTTGTTCTGACT.............................................................. | 52 | 1 | 41.00 | 41.00 | 41.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTGTTCTGACTTGTGTCTTTCAG.................................................. | 23 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CCAAAGATCCCGGACAAGCAGTTAGATG................ | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................AAACTATAACAGTTGTTCTGACTTGTGT......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ACAAGCAGTTAGATGAGAGGGAGCACA.... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AGATCCCGGACAAGCAGTTAGATGAGAG............ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CCAAAGATCCCGGACAAGCAGTTAGAT................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................TCCCGGACAAGCAGTTAGATGAGAGG........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCTGACTTGTGTCTTTCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CAAAGATCCCGGACAAGCAGTTAGATt................ | 27 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................CAAAGATCCCGGACAAGCAGTTAGAT................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................GATCCCGGACAAGCAGTTAGATGAGAGG........... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAACAGTTGTTCTGACTTGaaaa........................................................ | 23 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................CGGACAAGCAGTTAGATGAGAGGGAGC....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| AGAAAAAAGACAAACAGCTTATTACTTAGAAAGGAATCTTACAGAACTACTCTCCAGAAAGTTAAAGTTAATGATAACTGATTGACCTTTGTCTCATGATTCATTTTTTGCAAGTTGTATGTCTAGCTGATATTTCAAATAATTAATTGTATATATACATCCAGTAAACTATAACAGTTGTTCTGACTTGTGTCTTTCAGCCACCACCAAAGATCCCGGACAAGCAGTTAGATGAGAGGGAGCACACAAT ..........................................................................................................................................................(((((..(((..((((.....))))...)))...)))))......................................................... ...................................................................................................................................132.................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................CCACCAAAGATCCCG................................ | 15 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - |
| ....................................................................GATAACTGATTgttt....................................................................................................................................................................... | 15 | gttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................CTGATTGACCTTTaccc................................................................................................................................................................ | 17 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................TACAGAACTACTCTtgca..................................................................................................................................................................................................... | 18 | tgca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |