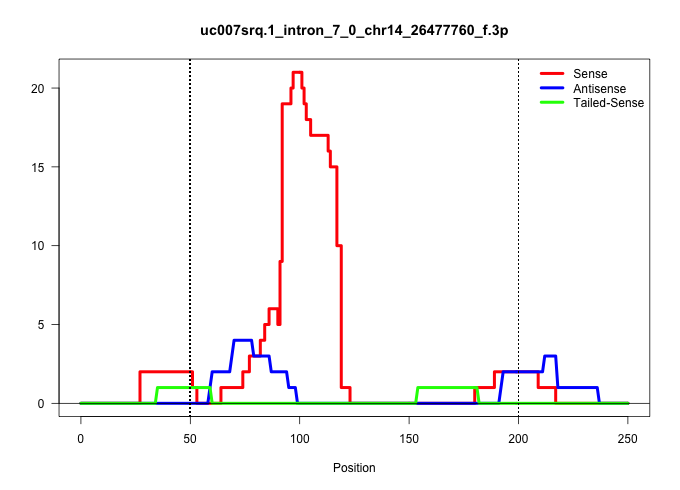
| Gene: Zmiz1 | ID: uc007srq.1_intron_7_0_chr14_26477760_f.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| TTGAGTGTGGGTATGCCTGCCCCCAGCTGAGATGCAGTGGTCTTGGGGACAGCCGGCAGGCCCATACTTGACACTGTTAAGTTTTCTGTCTTTGAGGTAGAACTCCAGGAGGCAAGCGGAAGGGAGTGAGAAGGCCAGAGAGAGCCCTCACTCCTCCTGGAGCAGCCTCAGACACCTCAACGAGGCTTCTTCTTTTCTAGCTACTGCCGGAACTCACAAACCCGGATGAGCTTCTTTCCTACCTGGACCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................TGAGGTAGAACTCCAGGAGGCAAGCGG................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | - | - | - | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - |
| ............................................................................................TGAGGTAGAACTCCAGGAGGCAAGC..................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTGAGGTAGAACTCCAGGAGGCAAGCGG................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................TGAGATGCAGTGGTCTTGGGGACA....................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TTTCTGTCTTTGAGGTAGA..................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................TAGAACTCCAGGAGGCAAGCGGAAGG............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................GTAGAACTCCAGGAGGCA........................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CGAGGCTTCTTCTTTTCTAGCTACTGCCG......................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................TACTTGACACTGTTAAGTTTTCTGTC................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................TGTTAAGTTTTCTGTCTTTGAGGTAGAA.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCCTGGAGCAGCCTCAGACACCTCAACa.................................................................... | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TAAGTTTTCTGTCTTTGAGGTAGAAC................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................AGTGGTCTTGGGGACAGCCGGCAGa.............................................................................................................................................................................................. | 25 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTCTTTTCTAGCTACTGCCGGAACTCAC................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTGTCTTTGAGGTAGAACTC................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGAGATGCAGTGGTCTTGGGGACAGC..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGTCTTTGAGGTAGAACTCCAGGAGGC......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| TTGAGTGTGGGTATGCCTGCCCCCAGCTGAGATGCAGTGGTCTTGGGGACAGCCGGCAGGCCCATACTTGACACTGTTAAGTTTTCTGTCTTTGAGGTAGAACTCCAGGAGGCAAGCGGAAGGGAGTGAGAAGGCCAGAGAGAGCCCTCACTCCTCCTGGAGCAGCCTCAGACACCTCAACGAGGCTTCTTCTTTTCTAGCTACTGCCGGAACTCACAAACCCGGATGAGCTTCTTTCCTACCTGGACCC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................ACACTGTTAAGTTTTCTGTCTTTGAGGTA....................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CTCACAAACCCGGATGAGCTTCTTT............. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCATACTTGACACTGTTAAGTTTTCT................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTTTCTAGCTACTGCCGGAACTCACA................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GCCCATACTTGACACTGTTA........................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................TTTCTAGCTACTGCCGGAACTCACA................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................GACACTGTTAAGTTTTCTGTCTTTGA........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |