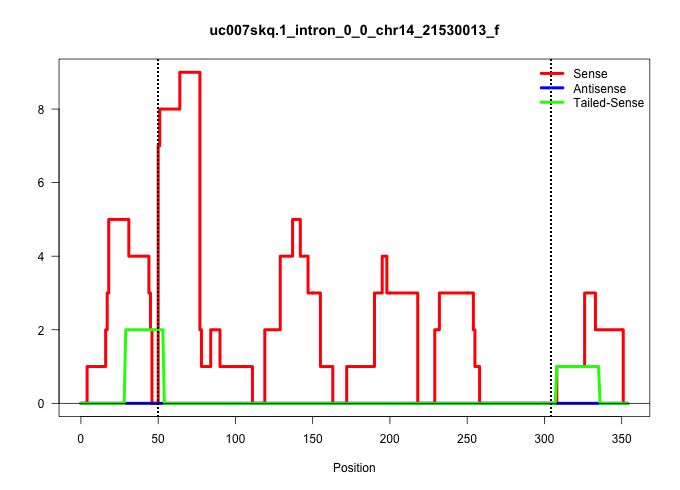
| Gene: 2310021P13Rik | ID: uc007skq.1_intron_0_0_chr14_21530013_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
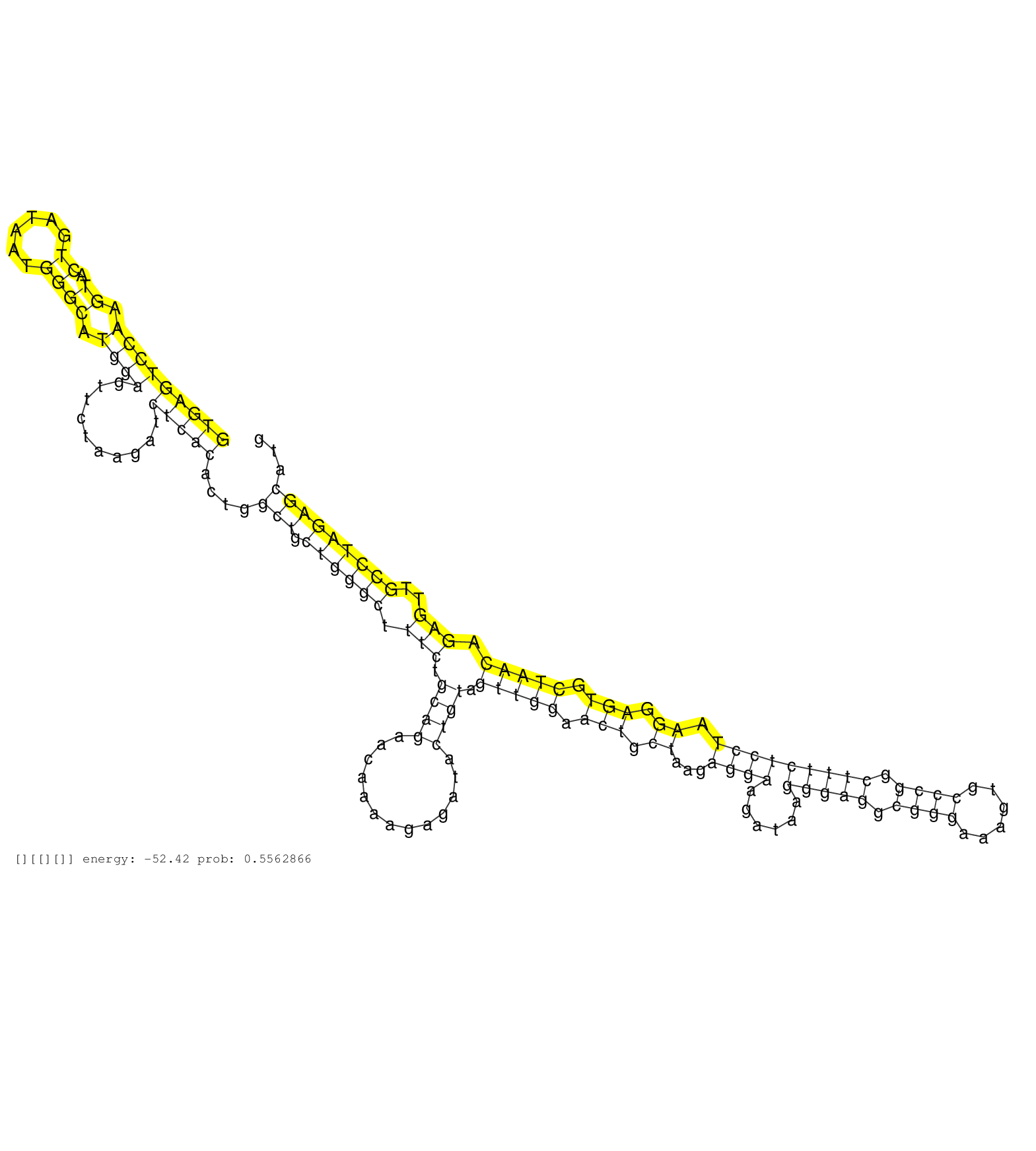
| CAACTCCGAATTGCTTTTTGGAGCTTCCCTGAGAATGAAGAGGACATTCGGTGAGTCCAAGTACTGATAATGGGCATGGAGTTCTAAGATCTCACACTGGCTGCTGGGCTTTCTGCAGAACAAAAGAGATACTGTAGTTGGAACTGCTAAGAGGAAGATAAGGGAGGCGGGAAAGTGCCCGGCTTTCTCCTAAGGAGTGCTAACAGAGTTGCCTAGAGCATGGTAGTCTCTCAGTGTCTGTGGAAGGTTATTTGCGGGGTTGTGGCCAAGTCCTCTTCTTCCCCCAGTCTGTATTCATGCCTAGCCAATGGCAGTGCGGATGAGTTTCAGCGAGGGGATCAGCTGTTCCGAATG ..................................................(((((((((.((.((......)))).))))..........)))))....(((.((((((.(((.((((.............)))).(((((.(((.((...((((......(((((.((((......)))).))))))))).)).))).))))).)))..)))))))))....................................................................................................................................... ..................................................51.........................................................................................................................................................................222.................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTCCAAGTACTGATAATGGGCAT..................................................................................................................................................................................................................................................................................... | 27 | 1 | 7.00 | 7.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................................................TAAGGAGTGCTAACAGAGTTGCCTAGAG........................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TACTGTAGTTGGAACTGCTAAGAGGA....................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................TCAGCGAGGGGATCAGCTGTTCCGA... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGAGAATGAAGAGGACATTCGtctg............................................................................................................................................................................................................................................................................................................ | 25 | tctg | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGGAGCTTCCCTGAGAATGAAGAGGACA.................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CTCAGTGTCTGTGGAAGGTTATTTGC................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................TAAGATCTCACACTGGCTGCTGGGCTT................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................TTGGAGCTTCCCTGAGAATGAAGAGGA...................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTCCAAGTACTGATAATGGGCATG.................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TTGGAACTGCTAAGAGGAAGATAAGG............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................AAGTGCCCGGCTTTCTCCTAAGGAGT............................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CTCAGTGTCTGTGGAAGGTTATTTG.................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................AGTGTCTGTGGAAGGTTATTTGCGGG................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................TGGCAGTGCGGATGAGTTTCAGCGAGGt.................. | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGGAACTGCTAAGAGGAAGATAAGGG.............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................TTTGGAGCTTCCCTGAGAATGAAGAGGAC..................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................TGGCAGTGCGGATGAGTTTCAGCGA..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................ACAAAAGAGATACTGTAGTTGGA.................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTAGTTGGAACTGCTAAGAGGAAG..................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................AGTGCTAACAGAGTTGCCTAGAG........................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................ACAAAAGAGATACTGTAGTTGGAACTGC............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................TGATAATGGGCATGGAGTTCTAAGAT........................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....TCCGAATTGCTTTTTGGAGCTTCCCTG................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAACTCCGAATTGCTTTTTGGAGCTTCCCTGAGAATGAAGAGGACATTCGGTGAGTCCAAGTACTGATAATGGGCATGGAGTTCTAAGATCTCACACTGGCTGCTGGGCTTTCTGCAGAACAAAAGAGATACTGTAGTTGGAACTGCTAAGAGGAAGATAAGGGAGGCGGGAAAGTGCCCGGCTTTCTCCTAAGGAGTGCTAACAGAGTTGCCTAGAGCATGGTAGTCTCTCAGTGTCTGTGGAAGGTTATTTGCGGGGTTGTGGCCAAGTCCTCTTCTTCCCCCAGTCTGTATTCATGCCTAGCCAATGGCAGTGCGGATGAGTTTCAGCGAGGGGATCAGCTGTTCCGAATG ..................................................(((((((((.((.((......)))).))))..........)))))....(((.((((((.(((.((((.............)))).(((((.(((.((...((((......(((((.((((......)))).))))))))).)).))).))))).)))..)))))))))....................................................................................................................................... ..................................................51.........................................................................................................................................................................222.................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................CTAACAGAGTTGCtat.............................................................................................................................................. | 16 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................ACTGCTAAGAGGAAGggt..................................................................................................................................................................................................... | 18 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |