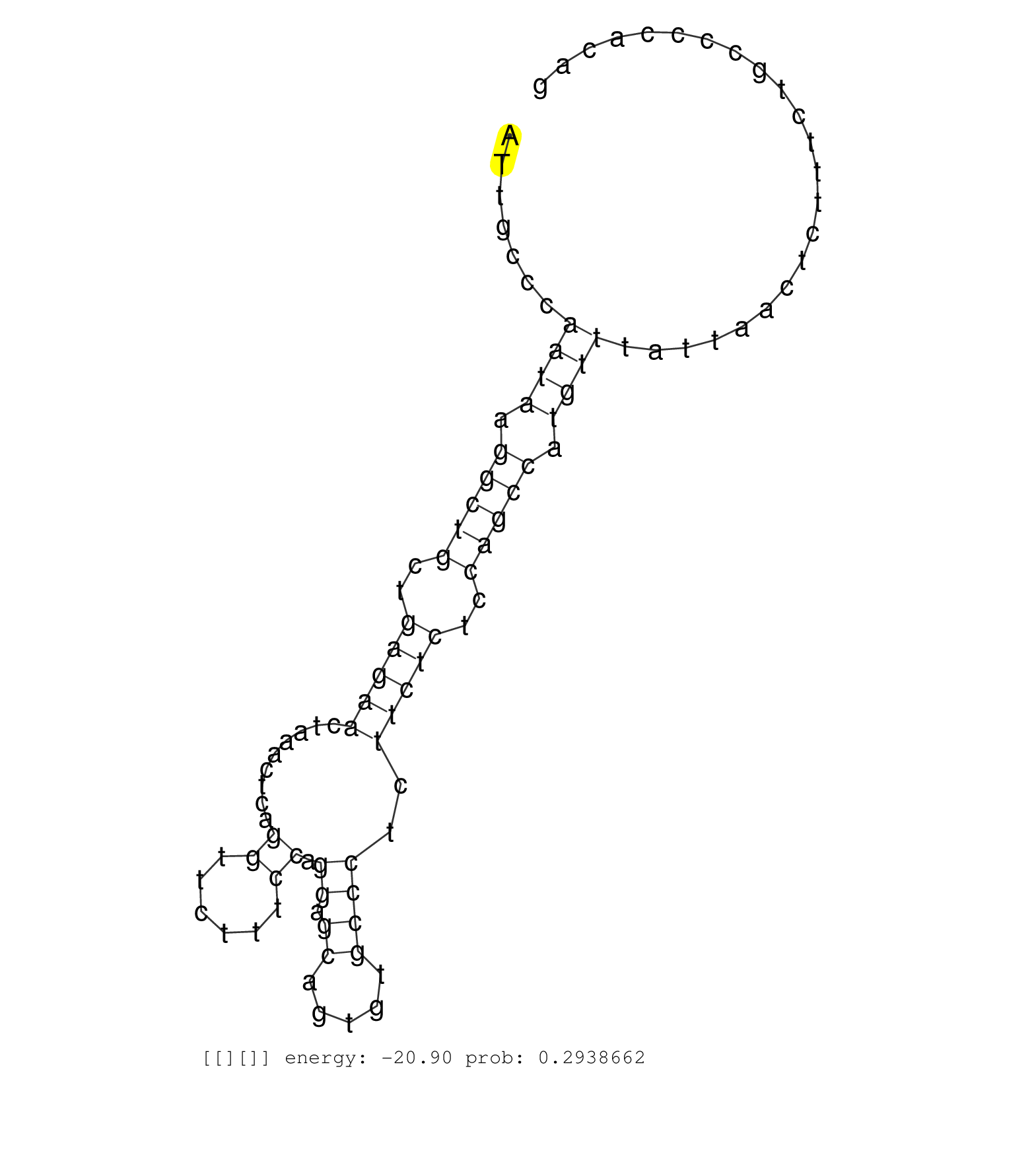
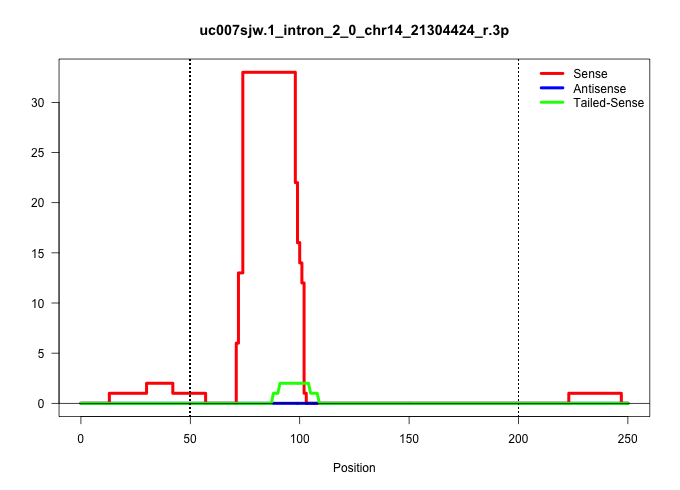
| Gene: Zmynd17 | ID: uc007sjw.1_intron_2_0_chr14_21304424_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(15) TESTES |
| TTATTTTTTAAAATTATGGGTTTTCACACATGAGATCAGTGCCTCCCAAGGCTAGAAGAGGACATGGGGTCTTCTAGAACTTGAATTACAGGAGATGTGAATTGCCCAATAAGGCTGCTGAGAACTAAACTCAGGTTCTTTCCAGGAGCAGTGTGCCCTCTTCTCTCCAGCCATGTTTATTAACTCTTTCTGCCCCACAGGTGATTTTGTCCTACCCTCAGGACCTTGGCCATGGCTACCTGAAGATATA ...........................................................................................................((((.(((((..(((((.........((......)).((.((.....))))..)))))..))))).))))......................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TAGAACTTGAATTACAGGAGATGTGAAT.................................................................................................................................................... | 28 | 1 | 11.00 | 11.00 | 1.00 | 3.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .......................................................................TTCTAGAACTTGAATTACAGGAGATGT........................................................................................................................................................ | 27 | 1 | 6.00 | 6.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TCTAGAACTTGAATTACAGGAGATGT........................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGAACTTGAATTACAGGAGATGTG....................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TCTAGAACTTGAATTACAGGAGATGTG....................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGAACTTGAATTACAGGAGATGTGA...................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TAGAACTTGAATTACAGGAGATGTGAA..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGAACTTGAATTACAGGAGATGTGAATT................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................GAGATGTGAATTGCagga............................................................................................................................................. | 18 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............TTATGGGTTTTCACACATGAGATCAGTGC................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CCTTGGCCATGGCTACCTGAAGAT... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................TAAACTCAGGTTCTTTCCAGGAGCAG................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................TGAGATCAGTGCCTCCCAAGGCTAGAA................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAGGAGATGTGAAcgta................................................................................................................................................. | 17 | cgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TTATTTTTTAAAATTATGGGTTTTCACACATGAGATCAGTGCCTCCCAAGGCTAGAAGAGGACATGGGGTCTTCTAGAACTTGAATTACAGGAGATGTGAATTGCCCAATAAGGCTGCTGAGAACTAAACTCAGGTTCTTTCCAGGAGCAGTGTGCCCTCTTCTCTCCAGCCATGTTTATTAACTCTTTCTGCCCCACAGGTGATTTTGTCCTACCCTCAGGACCTTGGCCATGGCTACCTGAAGATATA ...........................................................................................................((((.(((((..(((((.........((......)).((.((.....))))..)))))..))))).))))......................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................GGCTGCTGAGAACTAAACTggtt....................................................................................................................... | 23 | ggtt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCCAGCCATGTTTATTA.................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |