
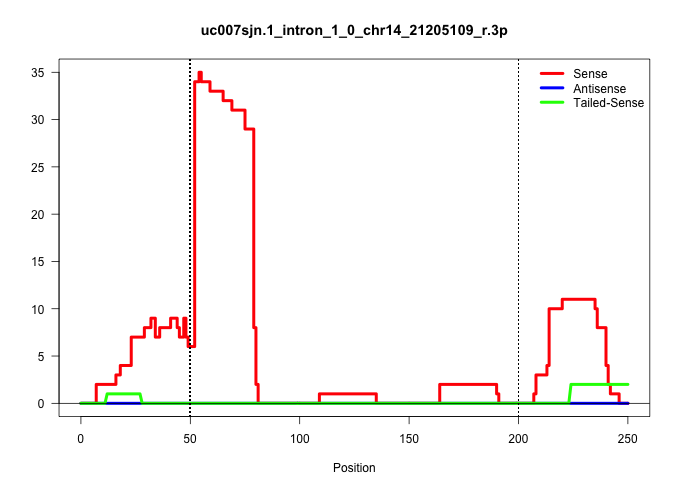
| Gene: Dnajc9 | ID: uc007sjn.1_intron_1_0_chr14_21205109_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| GAGATGCTGACGTACTTGTGCTCTAGATTTAGGAAGTAGAATGGGGATAACATAGAAGGACTTCCTGGAGTAAGACTGCTTCCATGAGCAGTTCCTAGACCTCCAGCTCTAGGAAGGGGCTAATGTTGTCTTTAAGGGGTAGAAAAAACGTATTTCTTACTTCTTGATTGCCATGGAGTCTGATAAAAGCTTCTGTCTAGGCTCAGGAAGAGGCTAAAGAAGCAGAGTTGAGCAGAAAGGAGCTGGGACT ......................................................((((..(((((((((((..((((((((....))))))))............))))))))))).((((.(((..((..(((((..(((((((.......))))).))..)))))..)))))..)))).........))))......................................................... ..................................................51............................................................................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................TAGAAGGACTTCCTGGAGTAAGACTGC........................................................................................................................................................................... | 27 | 1 | 21.00 | 21.00 | 9.00 | 5.00 | - | 4.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ....................................................TAGAAGGACTTCCTGGAGTAAGACTGCT.......................................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TAAAGAAGCAGAGTTGAGCAGAAAGG.......... | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................TAGAAGGACTTCCTGGAGTAAGACTGCTT......................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAACATAGAAGGACTTCCTGGAGTAAGA............................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGACGTACTTGTGCTCTAGATTTAGGA........................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................TAGATTTAGGAAGTAGAATGGGGAT.......................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGAGGCTAAAGAAGCAGAGTTGAGCAGA.............. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGATTGCCATGGAGTCTGATAAAAGCT........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CTAAAGAAGCAGAGTTGAGCAGAAAGGA......... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TAAAGAAGCAGAGTTGAGCAGAAAGGAG........ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAGTTGAGCAGAAAGGAGCTGGGACTgtat | 30 | gtat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............TACTTGTGCTCTAcct.............................................................................................................................................................................................................................. | 16 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AAGAGGCTAAAGAAGCAGAGTTGAGCAG............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................TGCTCTAGATTTAGGAAGTAGAATGGG............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGATTGCCATGGAGTCTGATAAAAGC............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAGTTGAGCAGAAAGGAGCTGGGACTa | 27 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TAGGAAGGGGCTAATGTTGTCTTTAA................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................TAGGAAGTAGAATGGGGATAACATAG................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGAATGGGGATAACATAGAAGGACTTCC......................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GAAGTAGAATGGGGATAACATAGAAGG............................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TAAAGAAGCAGAGTTGAGCAGAAAGGA......... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GAAGGACTTCCTGGAGTAAGACTGCT.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AGCAGAGTTGAGCAGAAAGGAGCTGG.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAGATTTAGGAAGTAGAATGGGGATA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGTGCTCTAGATTTAGGAAGTAGAATGG.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................TGGGGATAACATAGAAGGACTTCCTGGA..................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAGATGCTGACGTACTTGTGCTCTAGATTTAGGAAGTAGAATGGGGATAACATAGAAGGACTTCCTGGAGTAAGACTGCTTCCATGAGCAGTTCCTAGACCTCCAGCTCTAGGAAGGGGCTAATGTTGTCTTTAAGGGGTAGAAAAAACGTATTTCTTACTTCTTGATTGCCATGGAGTCTGATAAAAGCTTCTGTCTAGGCTCAGGAAGAGGCTAAAGAAGCAGAGTTGAGCAGAAAGGAGCTGGGACT ......................................................((((..(((((((((((..((((((((....))))))))............))))))))))).((((.(((..((..(((((..(((((((.......))))).))..)))))..)))))..)))).........))))......................................................... ..................................................51............................................................................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................GCTTCTGTCTAGGCTCAGGA.......................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |