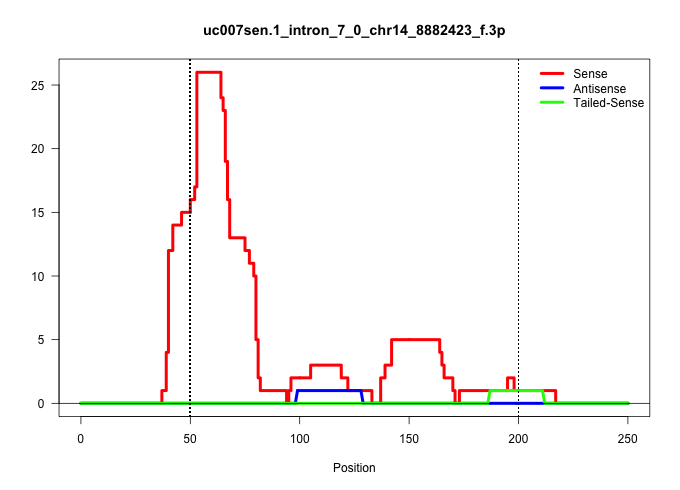
| Gene: Abhd6 | ID: uc007sen.1_intron_7_0_chr14_8882423_f.3p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| TACATTTGCGGGGGGGGGGGGGGGGGGGGCAAGCCCCTCTTGAGGAAGCAGAATGAACATCAGAATACAAACAGCCCCAGGGTGACCCACAACAATTGTGAGTCCTATGTGGGCAGAGCAGAAGCAGGAAGTCACACTGTACTTACAGAGATTAGAAATGCATGAAAAATATATGTTTCTTTCTTTCTCGTCCTCTGCAGGTGCTTGATGTGTCCGGGGCAGACATATTAGCCAAGTCAATCTCTAACTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................TGAACATCAGAATACAAACAGCCCCAG.......................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .......................................TTGAGGAAGCAGAATGAACATCAGAAT........................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGAACATCAGAATACAAACAGCCCCAGG......................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGGAAGCAGAATGAACATCAGAATA....................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................TGAGGAAGCAGAATGAACATCAGAA......................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGGAAGCAGAATGAACATCAGAATAC...................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................AGGAAGCAGAATGAACATCAGAATAC...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TACTTACAGAGATTAGAAATGCATGA..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTACAGAGATTAGAAATGCATGAAAAAT................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TCTTGAGGAAGCAGAATGAACATCAGA.......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................ATGAACATCAGAATACAAACAGCCCCAGGG........................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TATGTGGGCAGAGCAGAAGCAGGAAGTC..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGGTGCTTGATGTGTCCGG................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................AGCAGAATGAACATCAGAATACAAACAGC............................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................AGGAAGCAGAATGAACATCAGAATA....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGAGGAAGCAGAATGAACATCAGAAT........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TTGTGAGTCCTATGTGGGCAGAGC................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGTTTCTTTCTTTCTCGTCCTCTGC.................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................TACAAACAGCCCCAGGGTGACCCACAACA............................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GAATGAACATCAGAATACAAACAGCCC............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................TGAGGAAGCAGAATGAACATCAGA.......................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGTACTTACAGAGATTAGAAATGCATG...................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TCGTCCTCTGCAGGTGCTTGATGcg...................................... | 25 | cg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTACAGAGATTAGAAATGCATGAAAAATA............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTGAGTCCTATGTGGGCAGAGCAGA................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGAACATCAGAATACAAACAGCCCCA........................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................TGTACTTACAGAGATTAGAAATGCATGAA.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TACATTTGCGGGGGGGGGGGGGGGGGGGGCAAGCCCCTCTTGAGGAAGCAGAATGAACATCAGAATACAAACAGCCCCAGGGTGACCCACAACAATTGTGAGTCCTATGTGGGCAGAGCAGAAGCAGGAAGTCACACTGTACTTACAGAGATTAGAAATGCATGAAAAATATATGTTTCTTTCTTTCTCGTCCTCTGCAGGTGCTTGATGTGTCCGGGGCAGACATATTAGCCAAGTCAATCTCTAACTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................GAGTCCTATGTGGGCAGAGCAGAAGCAGGA......................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |