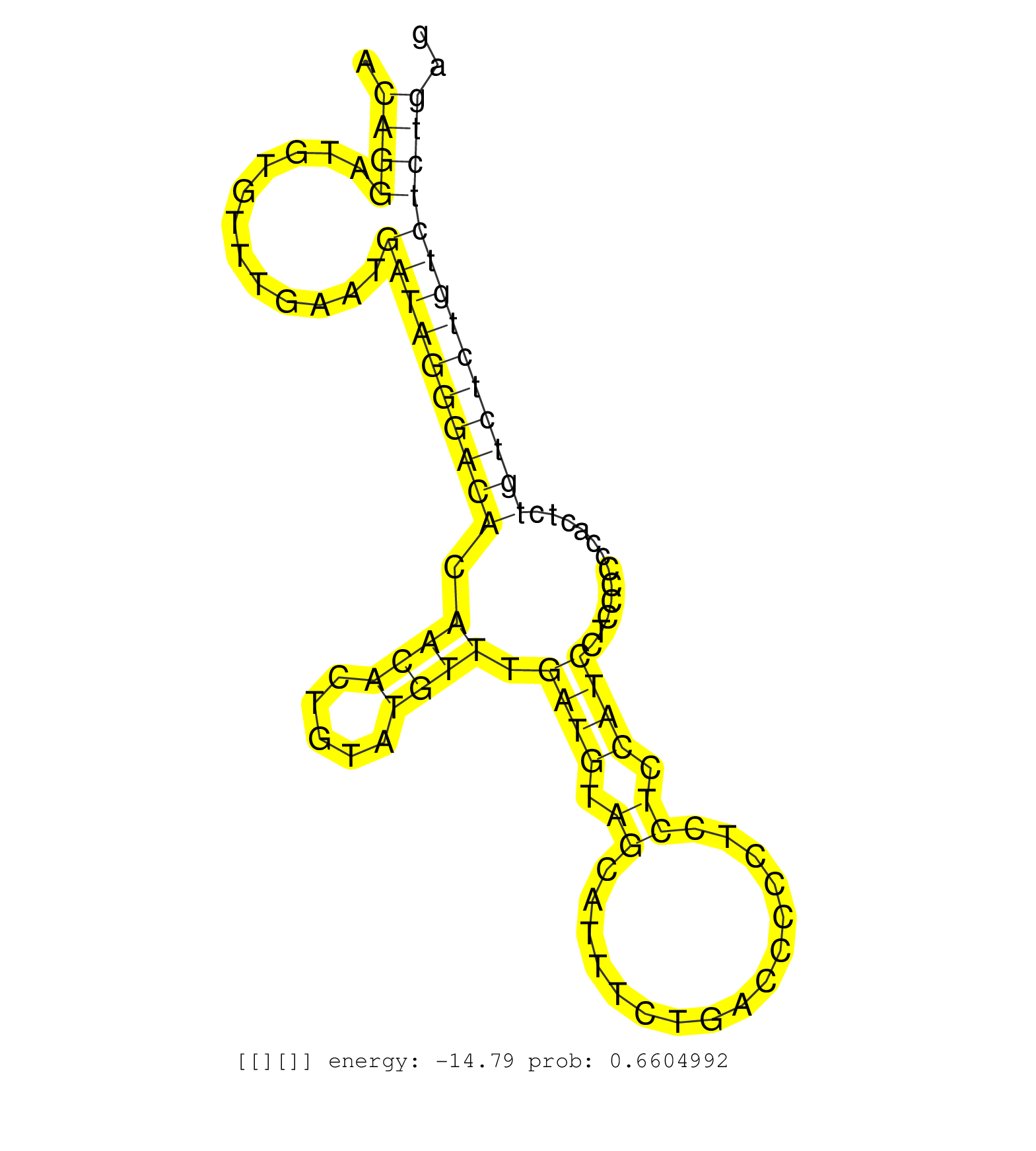

| Gene: Ipo11 | ID: uc007rtx.1_intron_18_0_chr13_107679487_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| ATTAACGGAAGAAGAACTGACCATGTGGGAGGAAGATCCAGAAGGCTTTAGTAAGGAAGAGTTTTTCAGTTTTCATATGACTTGTCTCTCTCTTTCATAAACAGGATGTGTTTGAATGATAGGGACACAACACTGTATGTTTGATGTAGCATTTCTGACCCCCTCCTCCATCCTCCCCCCACTCTGTCTCTGTCTCTGAGGCAGGTGTCACTGTGTAGCCCTGGCTGTCCTGACACTCACCATGTAGACC .....................................................................................................((((............((((((((((.((((.....)))).((((.((................)).))))............)))))))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................AAACAGGATGTGTTTGAATGATAGGGACACAACACTGTATGTTTGATGTAGC.................................................................................................... | 52 | 1 | 43.00 | 43.00 | 43.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGGATGTGTTTGAATGATAGGGACACAACACTGTATGTTTGATGTAGCATTT................................................................................................ | 52 | 1 | 38.00 | 38.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...AACGGAAGAAGAACTGACCATGTGGGAG........................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...AACGGAAGAAGAACTGACCATGTGGGA............................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................TGGGAGGAAGATCCAGAAGGCTTTAca...................................................................................................................................................................................................... | 27 | ca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TCACTGTGTAGCCCTGGCTGTCCaa.................. | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....ACGGAAGAAGAACTGACCATGTGGGAG........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...AACGGAAGAAGAACTGACCATGTGGGAGGA......................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TAACGGAAGAAGAACTGACCATGTGG.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....ACGGAAGAAGAACTGACCAT.................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................AGGATGTGTTTGAATtttt................................................................................................................................. | 19 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................CCATGTGGGAGGAAGATCCAGAAGGCTTTAcag..................................................................................................................................................................................................... | 33 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............AGAACTGACCATGTGGGAGGAAGATCCAG................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................CTGTGTAGCCCTGGCTGTCCTGAtaa.............. | 26 | taa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................GTTTGAATGATAGGGACACAA........................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................GTGTAGCCCTGGCTGTCCTGACctca............ | 26 | ctca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................TGTGGGAGGAAGATCCAGAAGGCTTT......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ACGGAAGAAGAACTGACCATGTGG.............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATTAACGGAAGAAGAACTGACCATGTGGGAGGAAGATCCAGAAGGCTTTAGTAAGGAAGAGTTTTTCAGTTTTCATATGACTTGTCTCTCTCTTTCATAAACAGGATGTGTTTGAATGATAGGGACACAACACTGTATGTTTGATGTAGCATTTCTGACCCCCTCCTCCATCCTCCCCCCACTCTGTCTCTGTCTCTGAGGCAGGTGTCACTGTGTAGCCCTGGCTGTCCTGACACTCACCATGTAGACC .....................................................................................................((((............((((((((((.((((.....)))).((((.((................)).))))............)))))))))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............TGACCATGTGGGAagc............................................................................................................................................................................................................................ | 16 | agc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CTGTGTAGCCCTGGCTGTCCTcgt................... | 24 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................GTGTAGCCCTGGCTGTCCTGcgt.................. | 23 | cgt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |