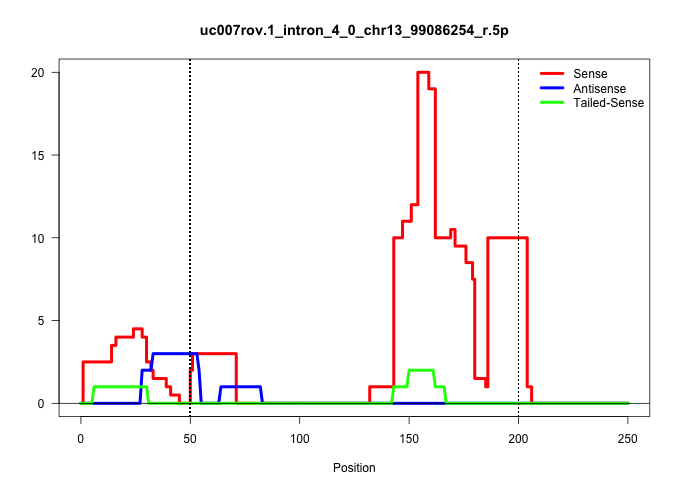
| Gene: Btf3 | ID: uc007rov.1_intron_4_0_chr13_99086254_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| CTGAGCGTGAACCTTAGTTCCCCGCGCGGCGCCATTTCCACTCCGACAAGGTACAGAAAAAAGGCCGTGGACGCCGGCGTGGGAGGAGACCGGGAGTTGGGGGTGGGGCGGGCGTTTCCCGGCAGGAGCGAGGCAGGGAGGGGACGGACGGAGCTGAGGAAAGCGATGCGACGGACGGGCGCTCCCACCCAGGCGGACTCTCGGGGGCGAGGGCGAGCCAGGGGCGGCTGGCCCGGGGCCGAAGCGACGC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................ACGGACGGAGCTGAGGAAA........................................................................................ | 19 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................ACCCAGGCGGACTCTCGG.............................................. | 18 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGAGGAAAGCGATGCGACGGACGGGC...................................................................... | 26 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..................................................GTACAGAAAAAAGGCCGTGGA................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGAGCGTGAACCTTAGTTCCCCGCGCGGC............................................................................................................................................................................................................................ | 29 | 2 | 1.50 | 1.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - |
| ......GTGAACCTTAGTTCCCCGCGCGGCa........................................................................................................................................................................................................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TACAGAAAAAAGGCCGTGGA................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GAGCTGAGGAAAGCGct................................................................................... | 17 | ct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACGGAGCTGAGGAAAGCGATGCGA............................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGAGGAAAGCGATGCGACGGACGGGCGCTC.................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGAGGAAAGCGATGCGACGGACGGG....................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CCACCCAGGCGGACTCTCGGGG............................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGCTGAGGAAAGCGATGCGACGGAC.......................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................ACGGACGGAGCTGAGGAga........................................................................................ | 19 | ga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................GCAGGGAGGGGACGGACGGAGCTGAGG........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................CGCGGCGCCATTTCCACTCCG............................................................................................................................................................................................................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .TGAGCGTGAACCTTAGTTCCCCGCGCGGCGC.......................................................................................................................................................................................................................... | 31 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..............TAGTTCCCCGCGCGGCGCCATTTCCAC................................................................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGAGCGTGAACCTTAGTTCCCCGCGCG.............................................................................................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GTTCCCCGCGCGGCGCC......................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TAGTTCCCCGCGCGGCGCCATTTCC................................................................................................................................................................................................................... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................GACGGACGGGCGCTCC................................................................. | 16 | 4 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGAGCGTGAACCTTAGTTCCCCGCGCGGCGCCATTTCCACTCCGACAAGGTACAGAAAAAAGGCCGTGGACGCCGGCGTGGGAGGAGACCGGGAGTTGGGGGTGGGGCGGGCGTTTCCCGGCAGGAGCGAGGCAGGGAGGGGACGGACGGAGCTGAGGAAAGCGATGCGACGGACGGGCGCTCCCACCCAGGCGGACTCTCGGGGGCGAGGGCGAGCCAGGGGCGGCTGGCCCGGGGCCGAAGCGACGC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................GCGCCATTTCCACTCCGACAAGGTACA................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ATTTCCACTCCGACAAGGTAC.................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CGGGCGTTTCCCgtcc.................................................................................................................................. | 16 | gtcc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCGTGGACGCCGGCGTGGG....................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |