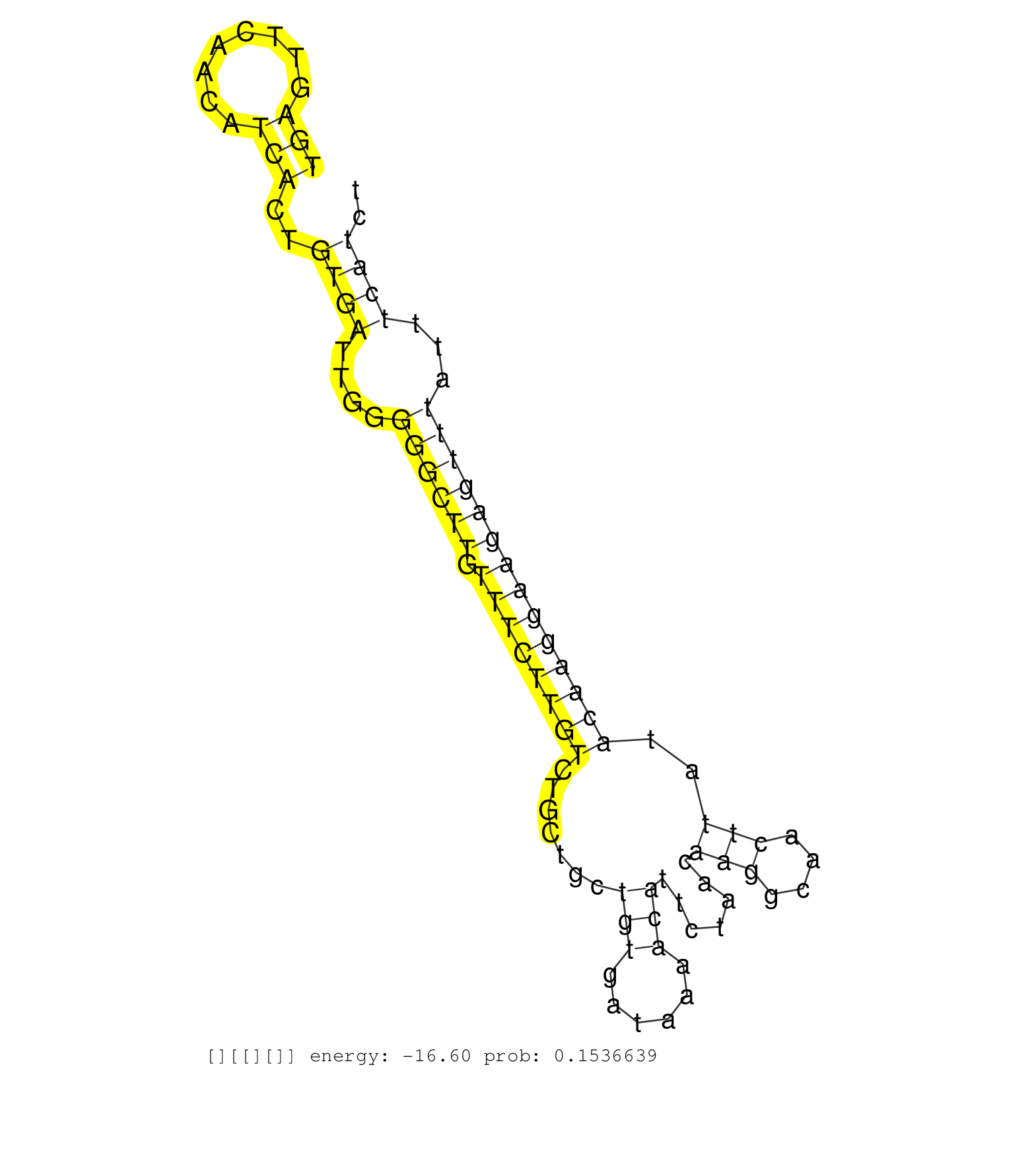
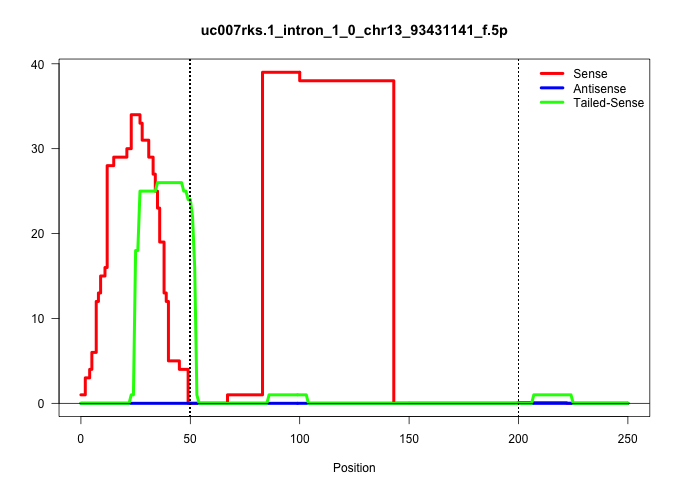
| Gene: Serinc5 | ID: uc007rks.1_intron_1_0_chr13_93431141_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(22) TESTES |
| TCTGCTGTGTGATGATGACACCCTCTGTGATGAAGCAGGTGAAGGACCACGTAAGTCTGTGGGTTTTGATAAGTTGTATGTGAAACTTGATGTAGAGGACTGAGTTCAACATCACTGTGATTGGGGGCTTGTTTCTTGTCTGCTGCTGTGATAAAACATTCTAACAAGGCAACTTATACAAGGAAGAGTTTATTTCATCTTATGGTTCCAGAGGGATAAGAGCCCATCACGGCGGCTTGGCATGACAGCA ....................................................................................................(((........)))..((((....((((((.((((((((.......(((......))).......(((....)))..))))))))))))))...)))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................AACTTGATGTAGAGGACTGAGTTCAACATCACTGTGATTGGGGGCTTGTTTC................................................................................................................... | 52 | 1 | 38.00 | 38.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGTGATGAAGCAGGTGAAGGACCACatt..................................................................................................................................................................................................... | 28 | att | 9.00 | 0.00 | - | 2.00 | 2.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGATGAAGCAGGTGAAGGACCACatt..................................................................................................................................................................................................... | 26 | att | 7.00 | 0.00 | - | 3.00 | 1.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGATGACACCCTCTGTGATGAAGCAGGT.................................................................................................................................................................................................................. | 28 | 1 | 7.00 | 7.00 | - | 1.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................TGTGATGAAGCAGGTGAAGGACCACat...................................................................................................................................................................................................... | 27 | at | 6.00 | 0.00 | - | 1.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGATGACACCCTCTGTGATGAAGCAG.................................................................................................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................TCTGTGATGAAGCAGGTGAAGGACCA......................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGTGATGAAGCAGGTGAAGGACCACa....................................................................................................................................................................................................... | 26 | a | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..TGCTGTGTGATGATGACACCCTCTGT.............................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .......TGTGATGATGACACCCTCTGTGATGAA........................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGTGATGATGACACCCTCTGTGATGAAG....................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGATGATGACACCCTCTGTGATGAAGC...................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTGTGATGAAGCAGGTGAAGG............................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CAGGTGAAGGACCACattc.................................................................................................................................................................................................... | 19 | attc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GATGACACCCTCTGTGATGAAGCAGGT.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CCTCTGTGATGAAGCAGGTGAAGGACCA......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................GTGAAACTTGATGTAGAGGA....................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GTGATGATGACACCCTCTGTGATGAAGC...................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCAGAGGGATAAGAGCat......................... | 18 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........ATGATGACACCCTCTGTGATGAAGCAG.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GATAAGTTGTATGTGAAACTTGATGTAGAGGAC...................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....CTGTGTGATGATGACACCCTCTGTGAT........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTGATGTAGAGGACgaca.................................................................................................................................................. | 18 | gaca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................TCTGTGATGAAGCAGGTGAAGGAt........................................................................................................................................................................................................... | 24 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGATGAAGCAGGTGAAGGACCACat...................................................................................................................................................................................................... | 25 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGTGATGAAGCAGGTGAAGGACaa......................................................................................................................................................................................................... | 24 | aa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGTGATGATGACACCCTCTGTGATGA......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGTGATGATGACACCCTCTGTGATGAAGC...................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGACACCCTCTGTGATGAAGCAGG................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGTGATGATGACACCCTCTGTGAT........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGTGATGATGACACCCTCTGTGATGA......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CCCTCTGTGATGAAGCA..................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TATGGTTCCAGAGGGATAAGAG............................ | 22 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 |
| TCTGCTGTGTGATGATGACACCCTCTGTGATGAAGCAGGTGAAGGACCACGTAAGTCTGTGGGTTTTGATAAGTTGTATGTGAAACTTGATGTAGAGGACTGAGTTCAACATCACTGTGATTGGGGGCTTGTTTCTTGTCTGCTGCTGTGATAAAACATTCTAACAAGGCAACTTATACAAGGAAGAGTTTATTTCATCTTATGGTTCCAGAGGGATAAGAGCCCATCACGGCGGCTTGGCATGACAGCA ....................................................................................................(((........)))..((((....((((((.((((((((.......(((......))).......(((....)))..))))))))))))))...)))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................GTTCAACATCACTgtct...................................................................................................................................... | 17 | gtct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |