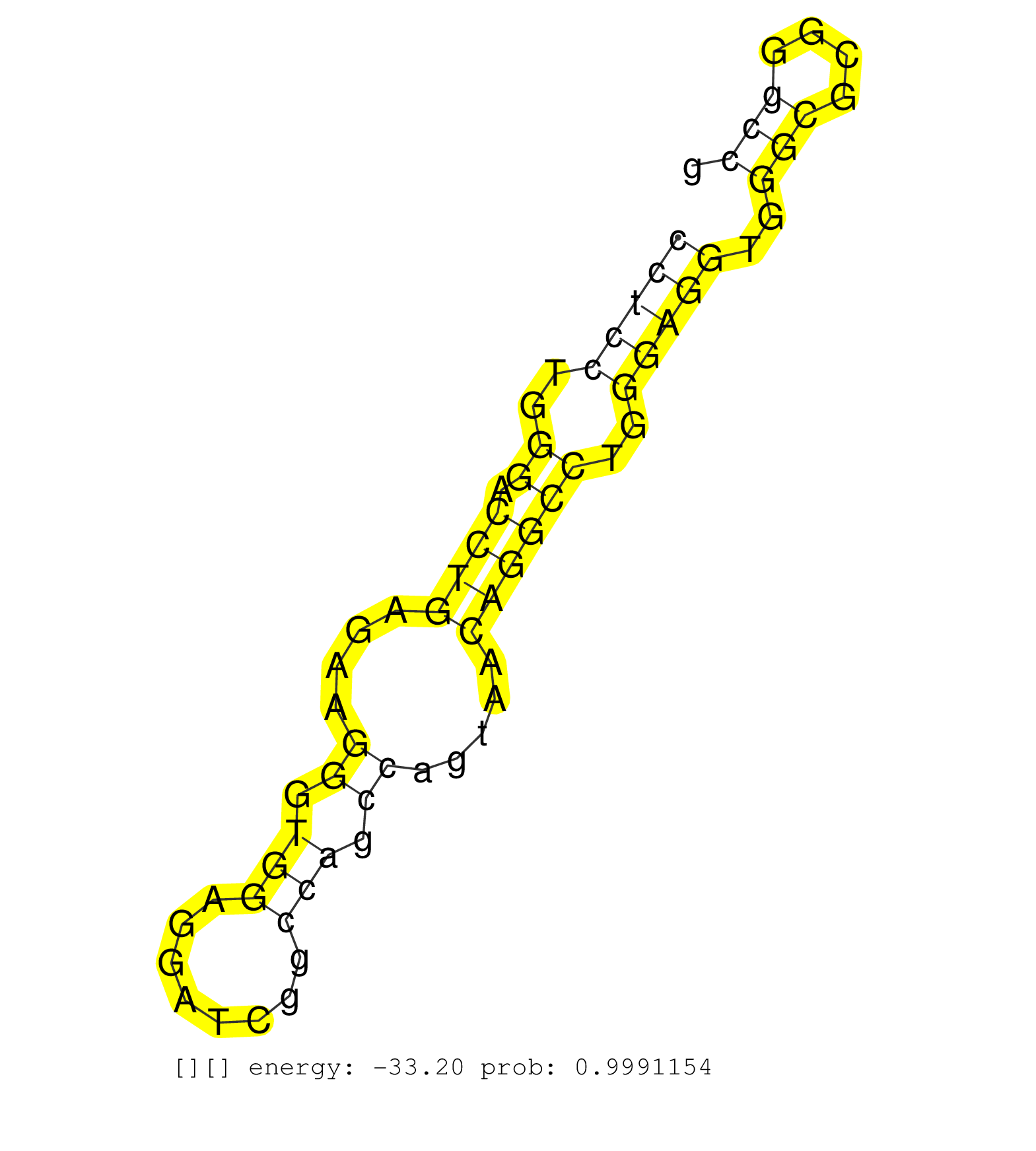
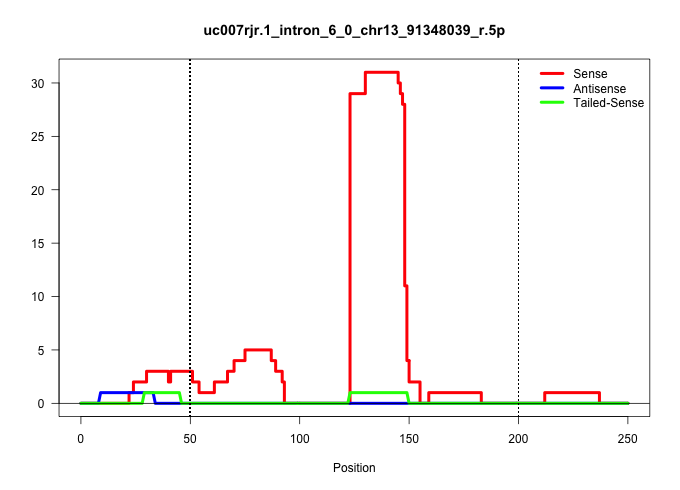
| Gene: Atg10 | ID: uc007rjr.1_intron_6_0_chr13_91348039_r.5p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CGCTCCGCCCCGTGCGCGCCGCCGGCTCCAGGTCAGGGCGAGCGAGCGGGGTGAGTACACCTAACCTCGGCGACCTAACCACGGCGGTACCTCCGCCGCTGCCTGCAGCCCTCGCCCGCCTCCTGGGACCTGAGAAGGGTGGAGGATCGGCCAGCCAGTAACAGGCCTGGGAGGTGGGCGCGGGCCGCCCAGGCGACCTTTCCACCTTGTGCTAGGAATGTGCTCACGTGACCTTGAAACAAAGGAAGTT ......................................................................................................................(((((..((.((((....((.(((........))).)).....))))))..)))))..(((....)))................................................................ ......................................................................................................................119.................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATC...................................................................................................... | 25 | 1 | 36.00 | 36.00 | 13.00 | 9.00 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCG..................................................................................................... | 26 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCGG.................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCGGC................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................TGAGAAGGGTGGAGGATCGGCCAGC............................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCGt.................................................................................................... | 27 | t | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGG......................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGttc...................................................................................................... | 25 | ttc | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CGACCTAACCACGGCGGTACCTC............................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GCTCCAGGTCAGGGCGAGCGAGCGGGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCGa.................................................................................................... | 27 | a | 1.00 | 7.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................ACCTCGGCGACCTAACCACGGCGGTACCT.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................TAACCACGGCGGTACCTC............................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TAACCTCGGCGACCTAACCACGGCGG................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGATCGGCCAG................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................GCGAGCGGGGTGAGTACACCTA........................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AACAGGCCTGGGAGGTGGGCGCGG................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................CGGCGACCTAACCACGGCGGTA................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................TTCCACCTTGTGCTAGGAATGTGCTC......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................CTCCAGGTCAGGGCGAGCGAGCGGGGT...................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GGTCAGGGCGAGCGAGCGGGGTGA.................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGA........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CGGCTCCAGGTCAGGGCG.................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................AGGTCAGGGCGAGCGtt............................................................................................................................................................................................................ | 17 | tt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGGGACCTGAGAAGGGTGGAGGAT....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TAGGAATGTGCTCACGTGACCTTGA............. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGCTCCGCCCCGTGCGCGCCGCCGGCTCCAGGTCAGGGCGAGCGAGCGGGGTGAGTACACCTAACCTCGGCGACCTAACCACGGCGGTACCTCCGCCGCTGCCTGCAGCCCTCGCCCGCCTCCTGGGACCTGAGAAGGGTGGAGGATCGGCCAGCCAGTAACAGGCCTGGGAGGTGGGCGCGGGCCGCCCAGGCGACCTTTCCACCTTGTGCTAGGAATGTGCTCACGTGACCTTGAAACAAAGGAAGTT ......................................................................................................................(((((..((.((((....((.(((........))).)).....))))))..)))))..(((....)))................................................................ ......................................................................................................................119.................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........CCGTGCGCGCCGCCGGCTCCAGGTC........................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |