
| Gene: Ccnh | ID: uc007riv.1_intron_2_0_chr13_85337167_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
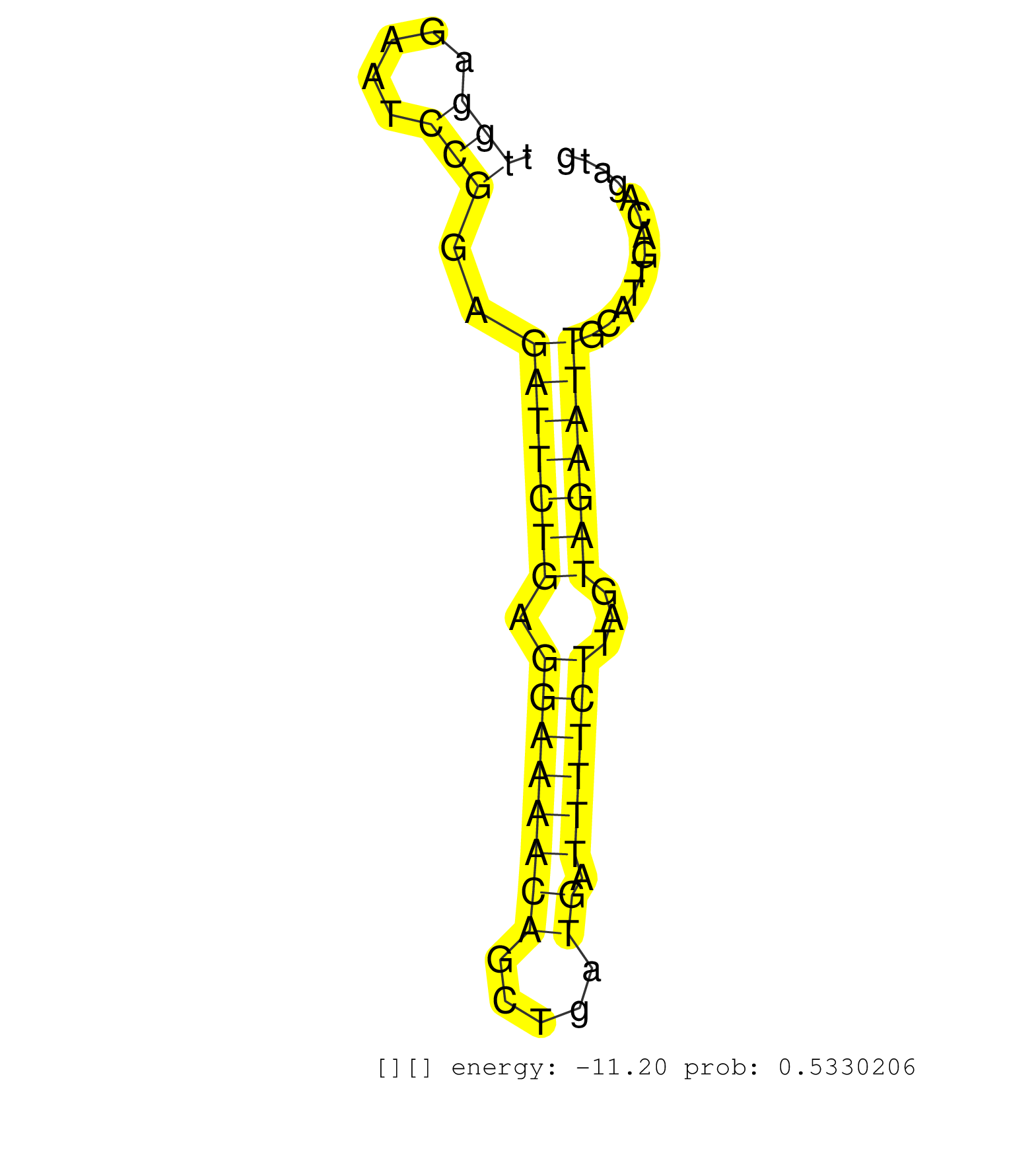
| TTCTTTATTAAGTACCTCCTAGTTACTGTAGTTTTTAATGGTGGCTAGCCAATAAACTCAGAAATGGTAACATCTATAAACTTTCTTTCTAGACTCGGTACCCCATGTTGGAGAATCCGGAGATTCTGAGGAAAACAGCTGATGATTTTCTTAGTAGAATTGCATTGACAGATGCTTATCTTTTATACACACCTTCACAGATTGCCCTGACCGCCATTTTATCAAGTGCCTCTAGAGCTGGAATTACTAT ............................................................................................................(((.....)))..(((((((.((((((((.....)).))))))...)))))))......................................................................................... ...........................................................................................................108...............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................TGATTTTCTTAGTAGAATTGCATTGACA................................................................................ | 28 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGATTTTCTTAGTAGAATTGCATTGAC................................................................................. | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................GAATCCGGAGATTCTGAGGAAAACAGCT.............................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCGGAGATTCTGAGGAAAACAGCTG............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGTTGGAGAATCCGGAGATTCTGAGG....................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................TAGTAGAATTGCATTGACAGATGCTTATC...................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................GAGAATCCGGAGATTCTGAGGAAAACAGC............................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................TCTTAGTAGAATTGCATTGACAGATGC........................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCCGGAGATTCTGAGGAAAACAGCTGA............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GGAGATTCTGAGGAAAACAGCTGAT........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................GAGATTCTGAGGAAAACAGCTGATGATT....................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................GAATCCGGAGATTCTGAGGAAAACAGC............................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................AATCCGGAGATTCTGAGGAAAACAGCTG............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................GTAGAATTGCATTGACAGATGCTT......................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................AGTAGAATTGCATTGACAGATGCTT......................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GATGATTTTCTTAGTAGAATTGCATT.................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................GATTGCCCTGACCGCCATTTTATC........................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCGGAGATTCTGAGGAAAACAGCT.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACAGCTGATGATTTTCTTAGTAGAATTGCAT..................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GGAGAATCCGGAGATTCTGAGGAAAACAGCT.............................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................TAGTAGAATTGCATTGACAGATGCTT......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AAAACAGCTGATGATTTTCTTActa.............................................................................................. | 25 | cta | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GGAGATTCTGAGGAAAACAGCTGA............................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................AATCCGGAGATTCTGAGGAAAACAGCT.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AAAACAGCTGATGATTTTCTTAGTA.............................................................................................. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TTTTATCAAGTGCCTCTAGAGCTGG......... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| TTCTTTATTAAGTACCTCCTAGTTACTGTAGTTTTTAATGGTGGCTAGCCAATAAACTCAGAAATGGTAACATCTATAAACTTTCTTTCTAGACTCGGTACCCCATGTTGGAGAATCCGGAGATTCTGAGGAAAACAGCTGATGATTTTCTTAGTAGAATTGCATTGACAGATGCTTATCTTTTATACACACCTTCACAGATTGCCCTGACCGCCATTTTATCAAGTGCCTCTAGAGCTGGAATTACTAT ............................................................................................................(((.....)))..(((((((.((((((((.....)).))))))...)))))))......................................................................................... ...........................................................................................................108...............................................................174.......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................GAGCTGGAATTACTActaa. | 19 | ctaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |