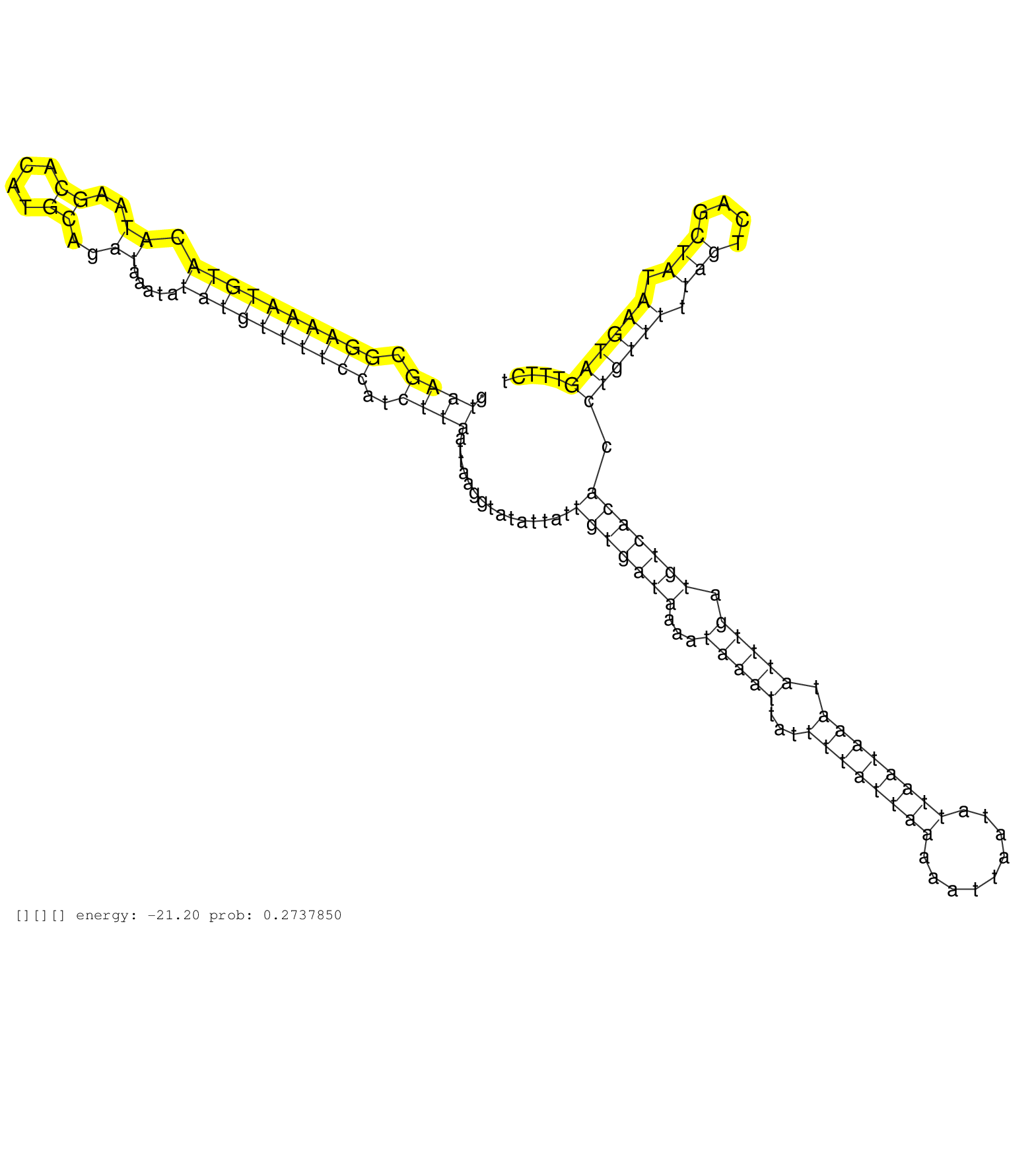
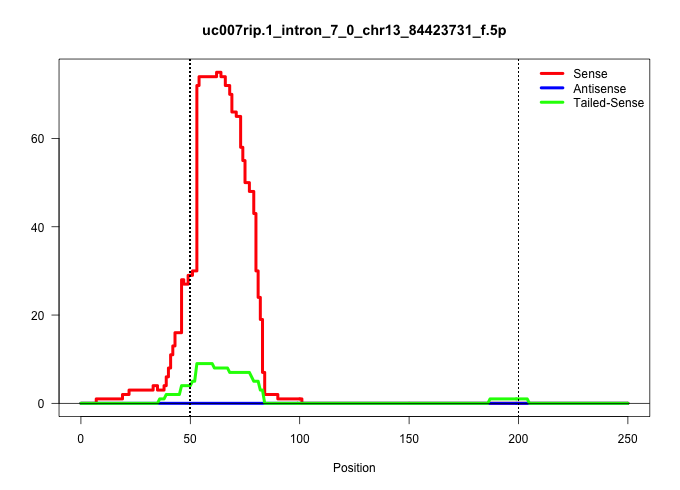
| Gene: Tmem161b | ID: uc007rip.1_intron_7_0_chr13_84423731_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(6) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
| GCTCAAATGCATCTGGATGCCCTGAATATGGCAACAGAAAAAATTACACAGTAAGCGGAAAATGTACATAAGCACATGCAGATAAATATATGTTTTCCATCTTAATTAAGGTATATTATTGTGATAAAATAAATTATTTTATTAAAAATTAATATTAATAAATATTTGATGTCACACCTGTTTTTAGTCAGCTATAAGTAGTTTCTCTTTGAAATTTCAGGCTAGTGTTAAAAATACATTTGTTGTTAAA ...................................................((((.((((((((((.((..((....))..)).....))))))))))..))))...............(((((((...(((((...((((((((.........)))))))).))))).))))))).((((((.(((....))).))))))................................................. ..................................................51.........................................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AGCGGAAAATGTACATAAGCACATGCA.......................................................................................................................................................................... | 27 | 1 | 13.00 | 13.00 | 3.00 | - | 1.00 | 4.00 | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAGAT....................................................................................................................................................................... | 30 | 1 | 11.00 | 11.00 | 1.00 | 5.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAG......................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAGA........................................................................................................................................................................ | 29 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGC................................................................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAGATA...................................................................................................................................................................... | 31 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGC........................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAGATt...................................................................................................................................................................... | 31 | t | 3.00 | 11.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGCA................................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGCAC............................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TTACACAGTAAGCGGAAAATGTACAT..................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................AGTAAGCGGAAAATGTACATAAGCAC............................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGCACAT............................................................................................................................................................................. | 31 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGCGGAAAATGTACATAAGCACATGC........................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCATCTGGATGCCCTGAATATGGCAAC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AATTACACAGTAAGCGGAAAATGTACA...................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................AAAAATTACACAGTAAGCGGAAAATGTACA...................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................ATTACACAGTAAGCGGAAAATGTACAT..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGCACATt............................................................................................................................................................................ | 32 | t | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................AGCACATGCAGATAAATATATGTTTTCCATC..................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AAATTACACAGTAAGCGGAAAATGTACATAA................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GCGGAAAATGTACATAAGCACATGCAGATA...................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCGGAAAATGTACATAAGCACATGCAGt........................................................................................................................................................................ | 29 | t | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AAAATTACACAGTAAGCGGAAAATGTACATA.................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TACACAGTAAGCGGAAAATGTACATAAGC................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................ATTACACAGTAAGCGGAAAATGTACATAAGC................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................GCGGAAAATGTACATAAGCACATGCAGAT....................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AAAATTACACAGTAAGCGGAAAATGTA........................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TCAGCTATAAGTAGTTgc............................................. | 18 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGCGGAAAATGTACATAAGCACATGCAGt........................................................................................................................................................................ | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................CACAGTAAGCGGAAAATGTACATAAGCACATGt........................................................................................................................................................................... | 33 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................AAAATTACACAGTAAGCGGAAAATGTACt...................................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AATTACACAGTAAGCGGAAAATGTACATAAGC................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................ACAGAAAAAATTACACAGTAAGCGGAAAATG.......................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TTACACAGTAAGCGGAAAATGTACATAAGC................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................TGAATATGGCAACAGAAAAAATTACACAG....................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................GAAAAAATTACACAGTAAGCGaatt............................................................................................................................................................................................. | 25 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................CCCTGAATATGGCAACAGAAAAAATTAC........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AATTACACAGTAAGCGGAAAATGTACAT..................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AAATTACACAGTAAGCGGAAAATGTA........................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................ATGGCAACAGAAAAAATTACAC......................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................TGTACATAAGCACATGCAGATAAATATA................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCTCAAATGCATCTGGATGCCCTGAATATGGCAACAGAAAAAATTACACAGTAAGCGGAAAATGTACATAAGCACATGCAGATAAATATATGTTTTCCATCTTAATTAAGGTATATTATTGTGATAAAATAAATTATTTTATTAAAAATTAATATTAATAAATATTTGATGTCACACCTGTTTTTAGTCAGCTATAAGTAGTTTCTCTTTGAAATTTCAGGCTAGTGTTAAAAATACATTTGTTGTTAAA ...................................................((((.((((((((((.((..((....))..)).....))))))))))..))))...............(((((((...(((((...((((((((.........)))))))).))))).))))))).((((((.(((....))).))))))................................................. ..................................................51.........................................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................ATTGTGATAAAATAggcc....................................................................................................................... | 18 | ggcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |