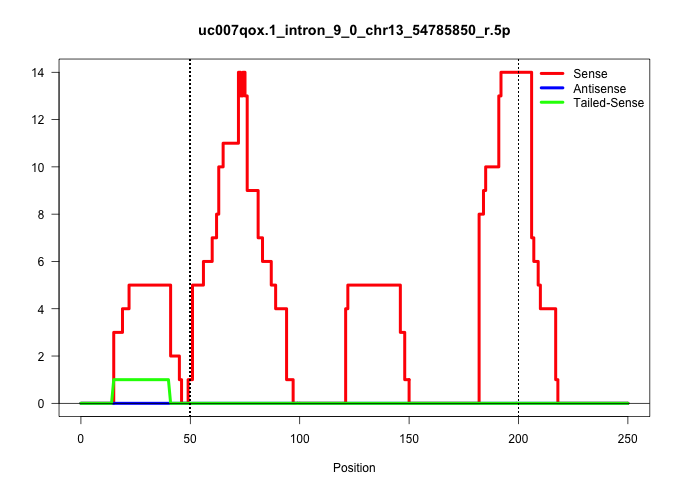
| Gene: Rnf44 | ID: uc007qox.1_intron_9_0_chr13_54785850_r.5p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(1) PIWI.mut |
(15) TESTES |
| GGAGCAACGCGCCGCTGCCTATTCGGGGTGGAGCCCAGCCGTGGCCCCAGGTAACGACCGGGCGCGGGCTGCTGTGGACGCGGCGGGAGGTGCGGGAGCTTCGGTGAGGCCTCGCCGGCCCTAGTCGTCACCTGGGCCGGTCTCTCGGTCCAAGTGCTAGGGCGCCGAGCTGGACGAGTTCCTGTGAGCTCTGCGCTCGGATGGTGGGACTCCAACGAGGATCCTGTGCCCTGTGGCCCCGTGAGGGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................TGTGAGCTCTGCGCTCGGATGGTG............................................ | 24 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAACGACCGGGCGCGGGCTGCTGTG.............................................................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTGGACGCGGCGGGAGGTGCG............................................................................................................................................................ | 22 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGCCTATTCGGGGTGGAGCCCAGCCG................................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................GCGGGCTGCTGTGGACGC......................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TAGTCGTCACCTGGGCCGGTCTCTC........................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGCGCTCGGATGGTGGGACTCCAACG................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TAGTCGTCACCTGGGCCGGTCTCTCGG...................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................AGTCGTCACCTGGGCCGGTCTCTCGGTC.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................TGGACGCGGCGGGAGGTGCGGGA......................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................TGAGCTCTGCGCTCGGATGGTGGGA......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................TATTCGGGGTGGAGCCCAGCCGTGGC............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................GGGCTGCTGTGGACGCGGCGGGAG................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGGGCTGCTGTGGACGCGG....................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................GCGCTCGGATGGTGGGACTCCAACG................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGTGAGCTCTGCGCTCGGATGGTGG........................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............TGCCTATTCGGGGTGGAGCCCAGCag................................................................................................................................................................................................................. | 26 | ag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTAACGACCGGGCGCGGGCTGCTGT............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGCGCTCGGATGGTGGGACTCCAACGA................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................TCGGGGTGGAGCCCAGCCGTGGCC............................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................GAGCTCTGCGCTCGGATGGTGGGAC........................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGCGGGCTGCTGTGGACGCGGCGGG................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................ACCGGGCGCGGGCTGCT................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GGAGCAACGCGCCGCTGCCTATTCGGGGTGGAGCCCAGCCGTGGCCCCAGGTAACGACCGGGCGCGGGCTGCTGTGGACGCGGCGGGAGGTGCGGGAGCTTCGGTGAGGCCTCGCCGGCCCTAGTCGTCACCTGGGCCGGTCTCTCGGTCCAAGTGCTAGGGCGCCGAGCTGGACGAGTTCCTGTGAGCTCTGCGCTCGGATGGTGGGACTCCAACGAGGATCCTGTGCCCTGTGGCCCCGTGAGGGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................GAGTTCCTGTGAGCTCtga........................................................... | 19 | tga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |