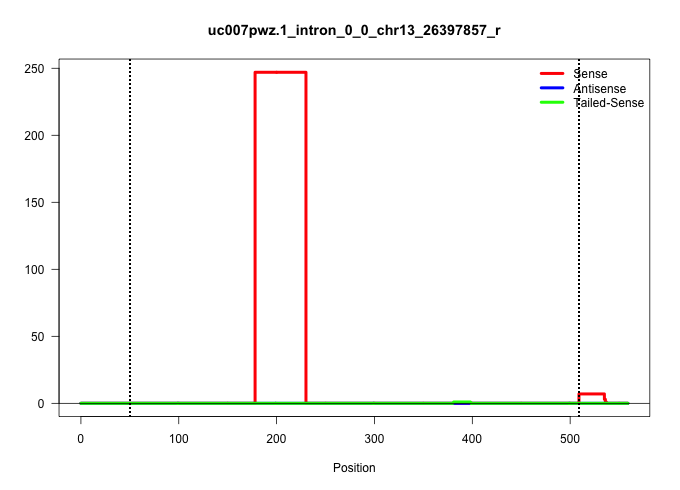
| Gene: AK007041 | ID: uc007pwz.1_intron_0_0_chr13_26397857_r | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.mut |
(7) TESTES |
| GGGTCACAGGAAGCTGATAGATTGAAAGGACATTGAAAATCCACAGGCAGGTAAGATACCCTTATTTACTCCATACTATAGCTGCTACATAGGAAGTTATTGATAAAGATCTAGGTATATGCTACACACAGGCAAGCATAAGCACACACATAGACTGGGCATATATAGGCTACATGCAACTTGTCATACAGACCTACAAACACAGAAGCAGGTTAGTGAAGGAAGCATCAAAGACAAAACACAAAAATGCCAAGACAAGGGCTCTGATCCCAACACCAGATTTATACAGAACCATACAGAAAGTCACATAAATGTTGAGCATAGTTGTGCAGCTGACTAGAGTGTTTAATTTTGGCACCTCTAGACATCTAATGTAGGCCATTTTTGCTGACACGTAGTTTATATATTTTTGGCATCAACAATGCCTTAGCTGACTCATACTTGATATCTCTATAGGCATCATGTTGTCTAAACATAAACTGGCCCTTAATGCCTTCTATTTCCAACAGTGAGGAATGGTGAGAAAGAAGTCATCTTCTGAAAGCCAGGTGTTCACCTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................ACTTGTCATACAGACCTACAAACACAGAAGCAGGTTAGTGAAGGAAGCATCA......................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 247.00 | 247.00 | 247.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGAGGAATGGTGAGAAAGAAGTCATC........................ | 26 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGAGGAATGGTGAGAAAGAAGTCATCT....................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TTTTTGCTGACACGatag................................................................................................................................................................ | 18 | atag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGAGGAATGGTGAGAAAGAAGTCATCTT...................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| GGGTCACAGGAAGCTGATAGATTGAAAGGACATTGAAAATCCACAGGCAGGTAAGATACCCTTATTTACTCCATACTATAGCTGCTACATAGGAAGTTATTGATAAAGATCTAGGTATATGCTACACACAGGCAAGCATAAGCACACACATAGACTGGGCATATATAGGCTACATGCAACTTGTCATACAGACCTACAAACACAGAAGCAGGTTAGTGAAGGAAGCATCAAAGACAAAACACAAAAATGCCAAGACAAGGGCTCTGATCCCAACACCAGATTTATACAGAACCATACAGAAAGTCACATAAATGTTGAGCATAGTTGTGCAGCTGACTAGAGTGTTTAATTTTGGCACCTCTAGACATCTAATGTAGGCCATTTTTGCTGACACGTAGTTTATATATTTTTGGCATCAACAATGCCTTAGCTGACTCATACTTGATATCTCTATAGGCATCATGTTGTCTAAACATAAACTGGCCCTTAATGCCTTCTATTTCCAACAGTGAGGAATGGTGAGAAAGAAGTCATCTTCTGAAAGCCAGGTGTTCACCTT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................ACAAAACACAAAAAaact........................................................................................................................................................................................................................................................................................................................ | 18 | aact | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................AGGCCATTTTTGCTcaga.......................................................................................................................................................................... | 18 | caga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 |