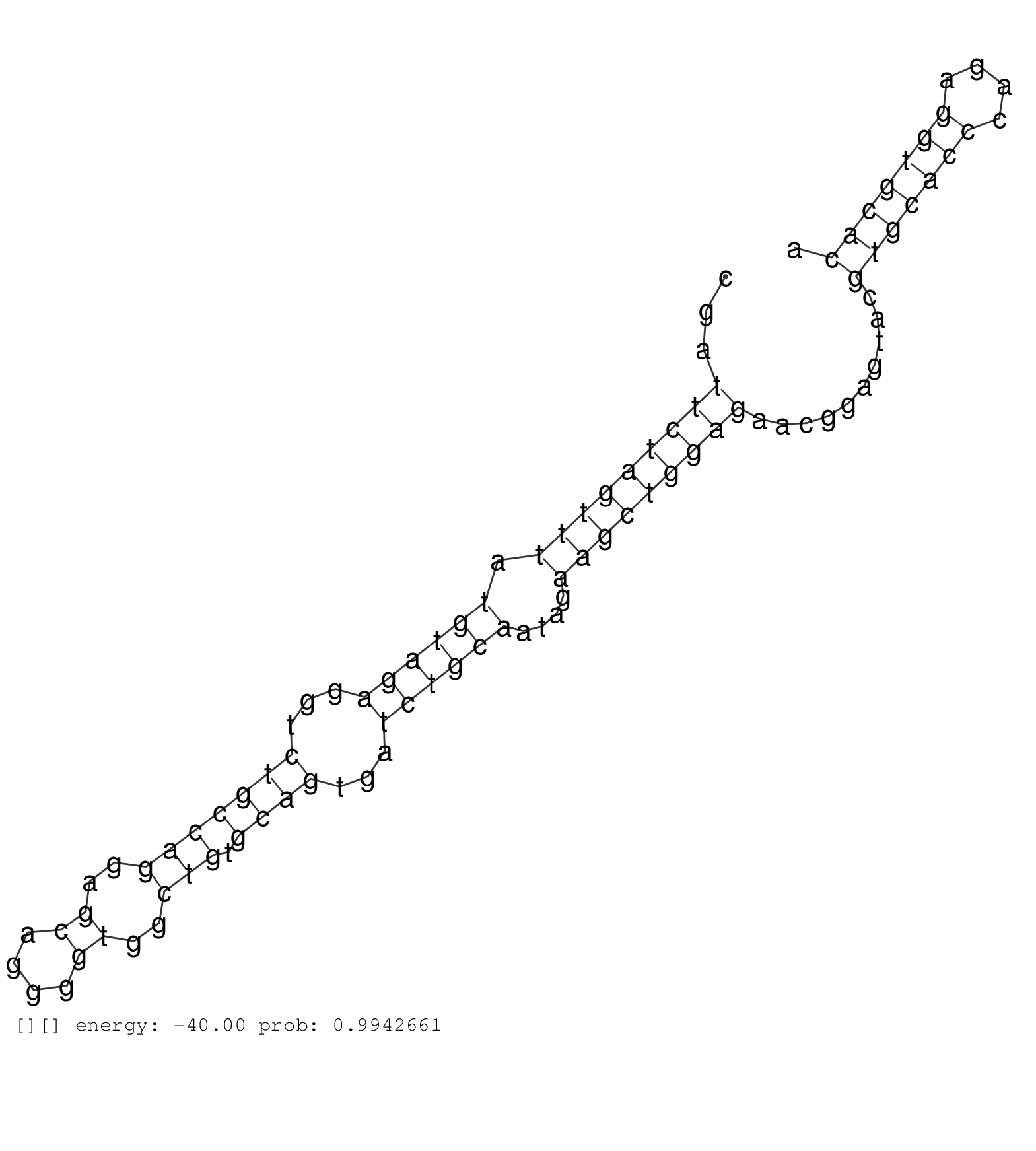

| Gene: Pgbd1 | ID: uc007pqk.1_intron_3_0_chr13_21526541_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(1) TDRD1.ip |
(16) TESTES |
| AATTTTAGCGTAGATCTGGGCAGGTTAGAGCCCTGGCGAAGGCACTACAGGTACATCTATTACGTTCGTCCGGTCTTTGTTGGGTATCAGGTACGTGCTGCGATTCTAGTTTATGTAGAGGTCTGCCAGGAGCAGGGGTGGCTGTGCAGTGATCTGCAATAGAAGCTGGAGAACGGAGTACGTGCACCCAGAGGTGCACACCCGGTTGCCACCTGTGTGGCTCAGTTTTCCCATTCCTATTTTAGGTTCG .......................................................................................................(((((((((.((((((...(((((((..((....))..))).))))...))))))....)))))))))..........(((((((....)))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGGGTA.................................................................................................................................................................... | 29 | 1 | 10.00 | 10.00 | 2.00 | 2.00 | 2.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGGGTAT................................................................................................................................................................... | 30 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGGGTATC.................................................................................................................................................................. | 31 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGG....................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ................TGGGCAGGTTAGAGCCCTGGCGAAGGCAC............................................................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGGG...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GCCCTGGCGAAGcgc.............................................................................................................................................................................................................. | 15 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................ATTACGTTCGTCCGGTCTTTGTTGGGTATC.................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGCGAAGGCACTACAGGTACATCTATTACt.......................................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TAGAGCCCTGGCGAAGGCACTAC.......................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................TATTACGTTCGTCCGGTCTTTGTTGGGaat................................................................................................................................................................... | 30 | aat | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGCGAAGGCACTACAGGTACATCTAT.............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................CGAAGGCACTACAGGTACATCTATT............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GTAGAGGTCTGCCAGGAG...................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................CGAAGGCACTACAGGTACATCTATTACGT......................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TACATCTATTACGTTCGTCCGGTCTT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................ACATCTATTACGTTCGTCCGGTCTTT............................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................CTATTACGTTCGTCCGGTCTTTGTTGGGTA.................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AATTTTAGCGTAGATCTGGGCAGGTTAGAGCCCTGGCGAAGGCACTACAGGTACATCTATTACGTTCGTCCGGTCTTTGTTGGGTATCAGGTACGTGCTGCGATTCTAGTTTATGTAGAGGTCTGCCAGGAGCAGGGGTGGCTGTGCAGTGATCTGCAATAGAAGCTGGAGAACGGAGTACGTGCACCCAGAGGTGCACACCCGGTTGCCACCTGTGTGGCTCAGTTTTCCCATTCCTATTTTAGGTTCG .......................................................................................................(((((((((.((((((...(((((((..((....))..))).))))...))))))....)))))))))..........(((((((....)))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................GAGCCCTGGCGAAGGCACTACAGGT...................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |