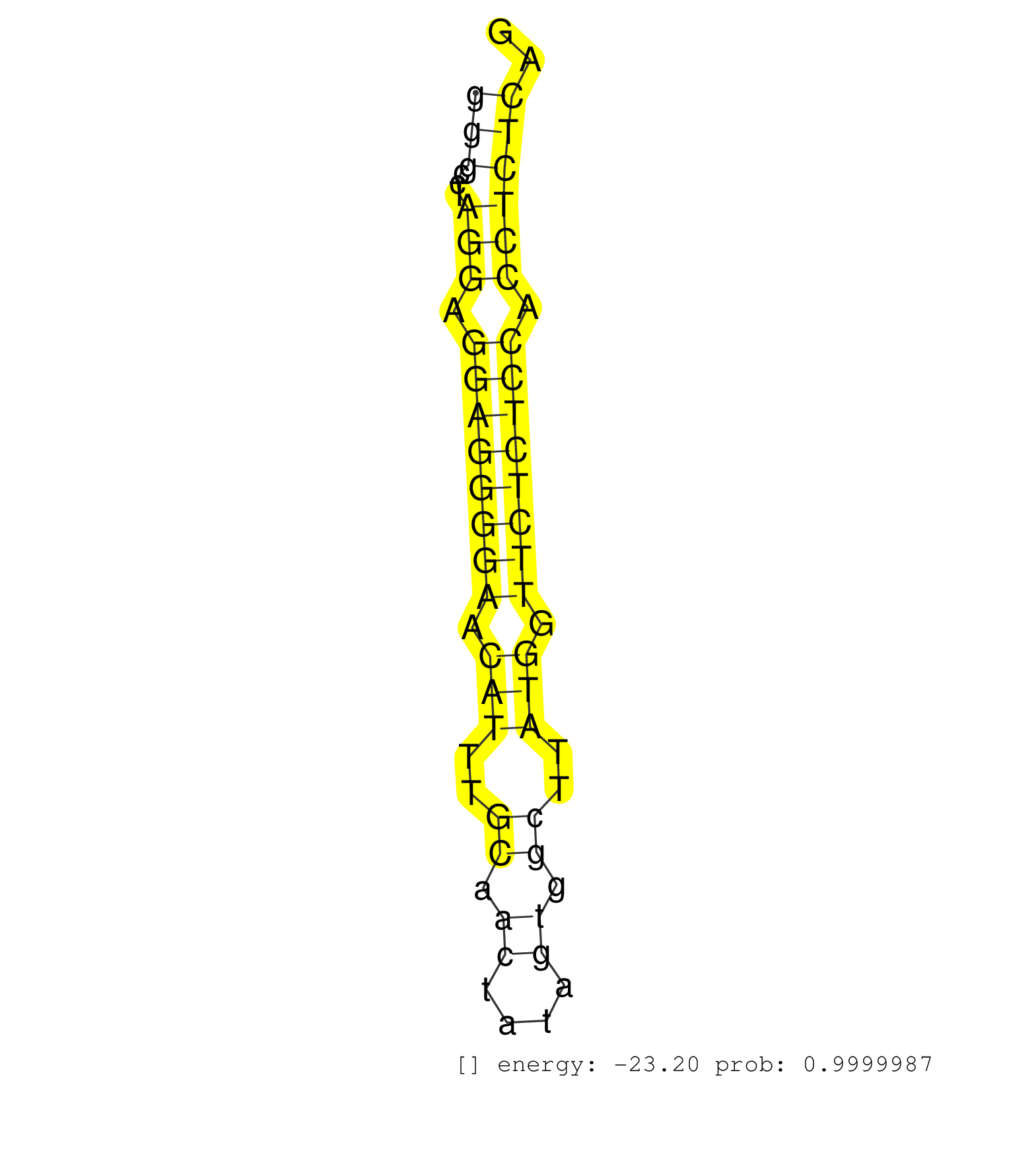

| Gene: Zkscan3 | ID: uc007pqi.1_intron_3_0_chr13_21488447_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TTAGGGTCTTCTGACTTACATTCAGTCTCTCCAGAACACCCTCACAGACATTTATGATTCTAAGTCCCACAAGATGACACAGAGACATTTGAACGTGGGGATCTCCTCAGCTGCTCTTTGGCAGTTTCCTTGGAGTCCTGGGCCTAGGAGGAGGGGAACATTTGCAACTATAGTGGCTTATGGTTCTCTCCACCTCTCAGTAGTCTTTTACAGAAATACAACTCACAGCTACAGTGAAACGGGAGAACTG ...........................................................................................................................................(((...(((.((((((((.(((..((.((....)).))..))).)))))))).)))))).................................................... ...........................................................................................................................................140.........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................TTTACAGAAATACAACTCACAGCTACAG................ | 28 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTG...................................................................................... | 20 | 1 | 6.00 | 6.00 | - | 4.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTGC..................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TACAGAAATACAACTCACAGCTACAGTGA............. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CTAGGAGGAGGGGAACATTTGaa.................................................................................... | 23 | aa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GGGGATCTCCTCAGCTG......................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTTTACAGAAATACAACTCACAGCTACA................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTGaa.................................................................................... | 22 | aa | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................CTAGGAGGAGGGGAACATTT....................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................CTAGGAGGAGGGGAACATTTGa..................................................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTATGGTTCTCTCCACCTCTCAat................................................. | 24 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................................TACAACTCACAGCTACAGTGAAACGG........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTGCAA................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TACAGAAATACAACTCACAGCTACAGTGAAA........... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CCTAGGAGGAGGGGAACATTT....................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GGGGATCTCCTCAGCTGCggc..................................................................................................................................... | 21 | ggc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TACAACTCACAGCTACAGTGAAACGGGA...... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................AAATACAACTCACAGCTACAGTGAAACGGGA...... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTGt..................................................................................... | 21 | t | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACATTTa...................................................................................... | 20 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGAGGAGGGGAACAT......................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TTAGGGTCTTCTGACTTACATTCAGTCTCTCCAGAACACCCTCACAGACATTTATGATTCTAAGTCCCACAAGATGACACAGAGACATTTGAACGTGGGGATCTCCTCAGCTGCTCTTTGGCAGTTTCCTTGGAGTCCTGGGCCTAGGAGGAGGGGAACATTTGCAACTATAGTGGCTTATGGTTCTCTCCACCTCTCAGTAGTCTTTTACAGAAATACAACTCACAGCTACAGTGAAACGGGAGAACTG ...........................................................................................................................................(((...(((.((((((((.(((..((.((....)).))..))).)))))))).)))))).................................................... ...........................................................................................................................................140.........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|