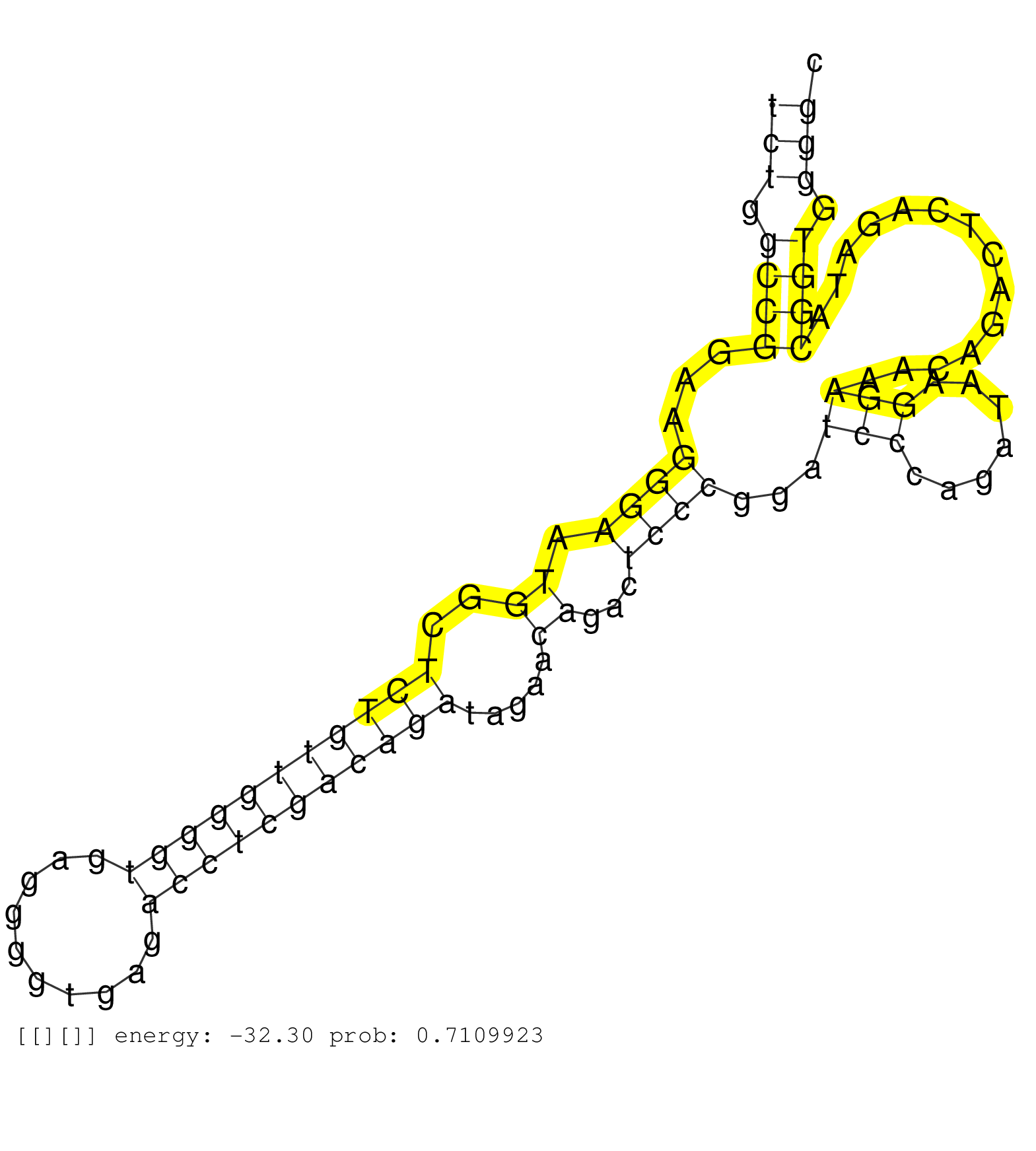
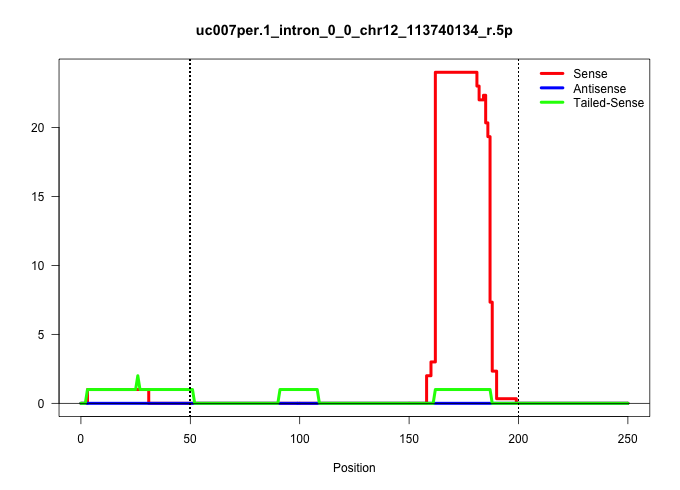
| Gene: Tmem179 | ID: uc007per.1_intron_0_0_chr12_113740134_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| AGCTGAACGTCGACAACTCTGCCTTCTACCATCAGTTTGCTATCGCCCAGGTAGGCGGGGTCTCGGGCAGGTAGGCGGGGTCTGTCTCTGGCCGGAAGGGAATGGCTCTGTTGGGGTGAGGGGTGAGACCTCGACAGATAGAACAGACTCCCGGATCCCAGATAAGGAAACAGACTCAGATACGGTGGGGCCATGCCGGGGAGGTGCAGGCTGTGGAGCAAGAGATTTCCTAGGAGAGGTTAGGAGAGTG ......................................................................................(((.((((...((((.((..(((((((((((..........))))))))))).....))...))))...(((.......)))..............)))).)))............................................................ ......................................................................................87......................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGTG............................................................... | 25 | 1 | 53.00 | 53.00 | 18.00 | 7.00 | 8.00 | 4.00 | 6.00 | - | 4.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGTGG.............................................................. | 26 | 1 | 19.00 | 19.00 | - | 4.00 | - | 3.00 | 1.00 | 5.00 | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................CAGATAAGGAAACAGACTCAGATACGG................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GATAAGGAAACAGACTCAGATACGGTG............................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGTGGGG............................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGgg............................................................... | 25 | gg | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AAGGAAACAGACTCAGATACGGTGGG............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGAT..................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................AAGGAAACAGACTCAGATACGGTGG.............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAACGTCGACAACTCTGCCTTCTACCA........................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGTGGG............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGG................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGcgg.............................................................. | 26 | cgg | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................AGGAAACAGACTCAGATACGGTGGGGC........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATA.................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAACGTCGACAACTCTGCCTTCa............................................................................................................................................................................................................................... | 24 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TACCATCAGTTTGCTATCGCCCAGtt...................................................................................................................................................................................................... | 26 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................CCGGAAGGGAATGGttat............................................................................................................................................. | 18 | ttat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................TGCAGGCTGTGGAGCAAGAGATTTCC.................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TAAGGAAACAGACTCAGATACGGT................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GGTGCAGGCTGTGGAG................................ | 16 | 9 | 0.78 | 0.78 | - | - | - | - | - | 0.78 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GTGGGGCCATGCCGG................................................... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| AGCTGAACGTCGACAACTCTGCCTTCTACCATCAGTTTGCTATCGCCCAGGTAGGCGGGGTCTCGGGCAGGTAGGCGGGGTCTGTCTCTGGCCGGAAGGGAATGGCTCTGTTGGGGTGAGGGGTGAGACCTCGACAGATAGAACAGACTCCCGGATCCCAGATAAGGAAACAGACTCAGATACGGTGGGGCCATGCCGGGGAGGTGCAGGCTGTGGAGCAAGAGATTTCCTAGGAGAGGTTAGGAGAGTG ......................................................................................(((.((((...((((.((..(((((((((((..........))))))))))).....))...))))...(((.......)))..............)))).)))............................................................ ......................................................................................87......................................................................................................191......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|