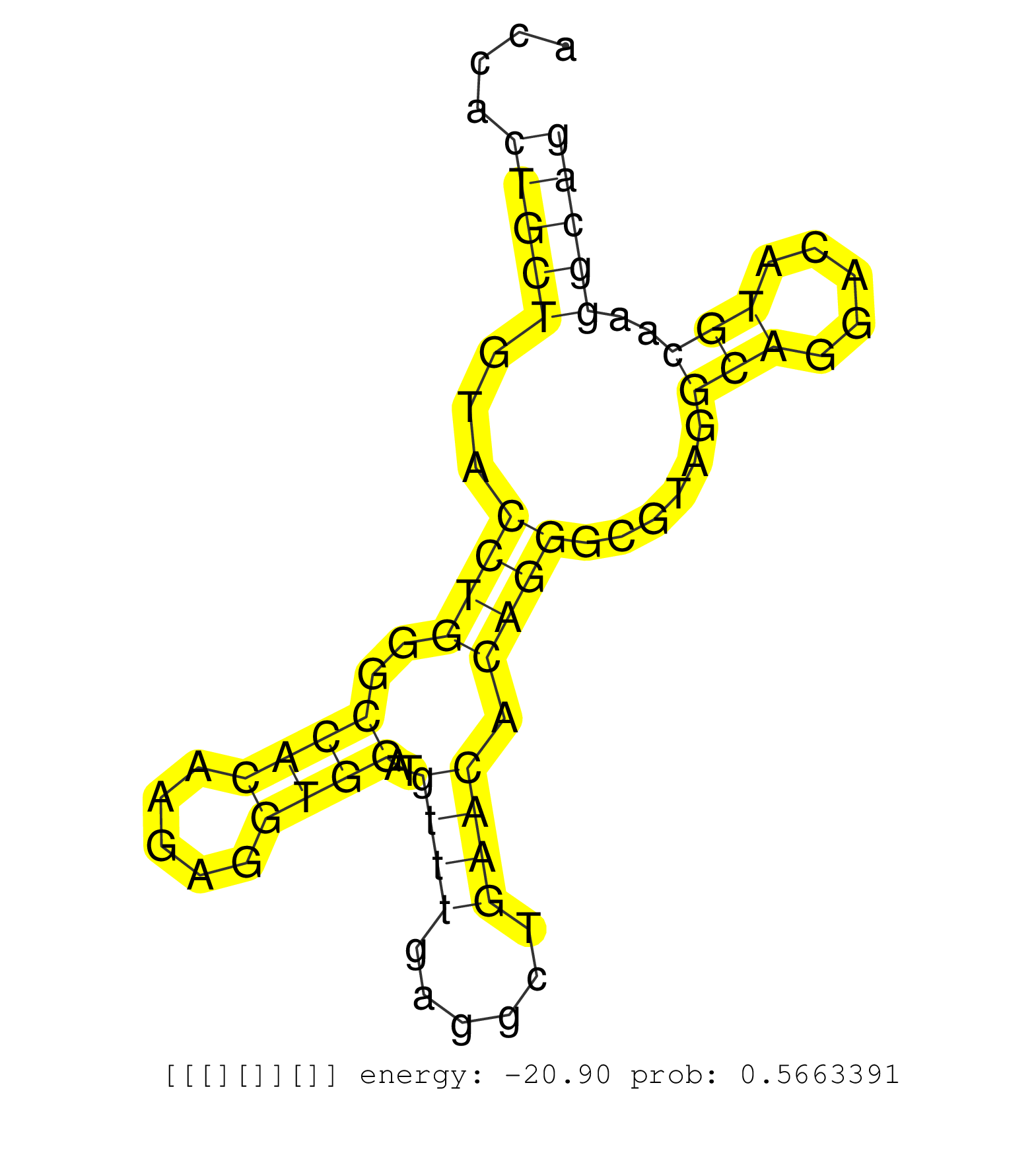

| Gene: BC115444 | ID: uc007pen.1_intron_0_0_chr12_113389581_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| GGAGCCAGAGTCAGCTGAATATCTGGTGGCCCTGGAGCAAGCCACAGCTGCCCTGGAGCAGTGCGTTAACCTGTGCAAGGCCCATGTCATGATGGTCACCTGCTTTGACATTGGAGTTGCTGCCACCACTGCTGTACCTGGGCCACAAGAGGTGGATGTTTGAGGCTGAACACAGGGCGTAGGCAGGACATGCAAGGCAGAGGGTGTGAGCGTAATGTGGGATGGAGTGAGGATGTGGAGGGGGTTGGCG ................................................................................................................................(((((...((((..((((.....))))..((((......)))).))))......(((.....)))..))))).................................................. ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TGCTGTACCTGGGCCACAAGAGGTGGAT............................................................................................. | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGCTGTACCTGGGCCACAAGAGGTGG............................................................................................... | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGCTGTACCTGGGCCACAAGAGGTGGA.............................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGTTTGAGGCTGAACACAGGGCGTAGGC.................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CACTGCTGTACCTGGGCCACAAG..................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GCTGAACACAGGGCGTAGGCA................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................AGGCTGAACACAGGGCGTAGGCAGGA.............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGAACACAGGGCGTAGGCAGGACATG.......................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TAATGTGGGATGGAGTGAGGATGTG............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................TGCTGTACCTGGGCCtgt....................................................................................................... | 18 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................GAGGTGGATGTTTGAGGCTGAACAC............................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GATGTTTGAGGCTGAACACAGGGCGT...................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................CATGCAAGGCAGAGGGTGTGAGCGTA.................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AGGTGGATGTTTGAGGCTGAACACAGGGC........................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGAGGCTGAACACAGGGCGTAGGCA................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................AGTGCGTTAACCTGTG............................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GCTGAACACAGGGCGTAGGCAGGACATGC......................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................GGATGTTTGAGGCgc.................................................................................. | 15 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................TGAGGCTGAACACAGGGCGTAGGCAGt............................................................... | 27 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CCACTGCTGTACCTGGGCCACAAGAGG.................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CTGCTGTACCTGGGCCACAAGAGGTG................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................TGAGCGTAATGTGGGATGGAGTGAG................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CACTGCTGTACCTGGGCCACAAGAGGTGGA.............................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGTTTGAGGCTGAACACAGGGCGTAGG................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................ACACAGGGCGTAGGCAGGACATGCAA....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................ACATGCAAGGCAGAGGGTGTGAGCGT..................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGCTGTACCTGGGCCACAAGAGGTGGATc............................................................................................ | 29 | c | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................CGTAATGTGGGATGGAGTGAGGATGT.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................TGAGGCTGAACACAGGGCGTAGGCAG................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGCAAGGCAGAGGGTGTGAGCGTAA................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAGCCAGAGTCAGCTGAATATCTGGTGGCCCTGGAGCAAGCCACAGCTGCCCTGGAGCAGTGCGTTAACCTGTGCAAGGCCCATGTCATGATGGTCACCTGCTTTGACATTGGAGTTGCTGCCACCACTGCTGTACCTGGGCCACAAGAGGTGGATGTTTGAGGCTGAACACAGGGCGTAGGCAGGACATGCAAGGCAGAGGGTGTGAGCGTAATGTGGGATGGAGTGAGGATGTGGAGGGGGTTGGCG ................................................................................................................................(((((...((((..((((.....))))..((((......)))).))))......(((.....)))..))))).................................................. ............................................................................................................................125........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|