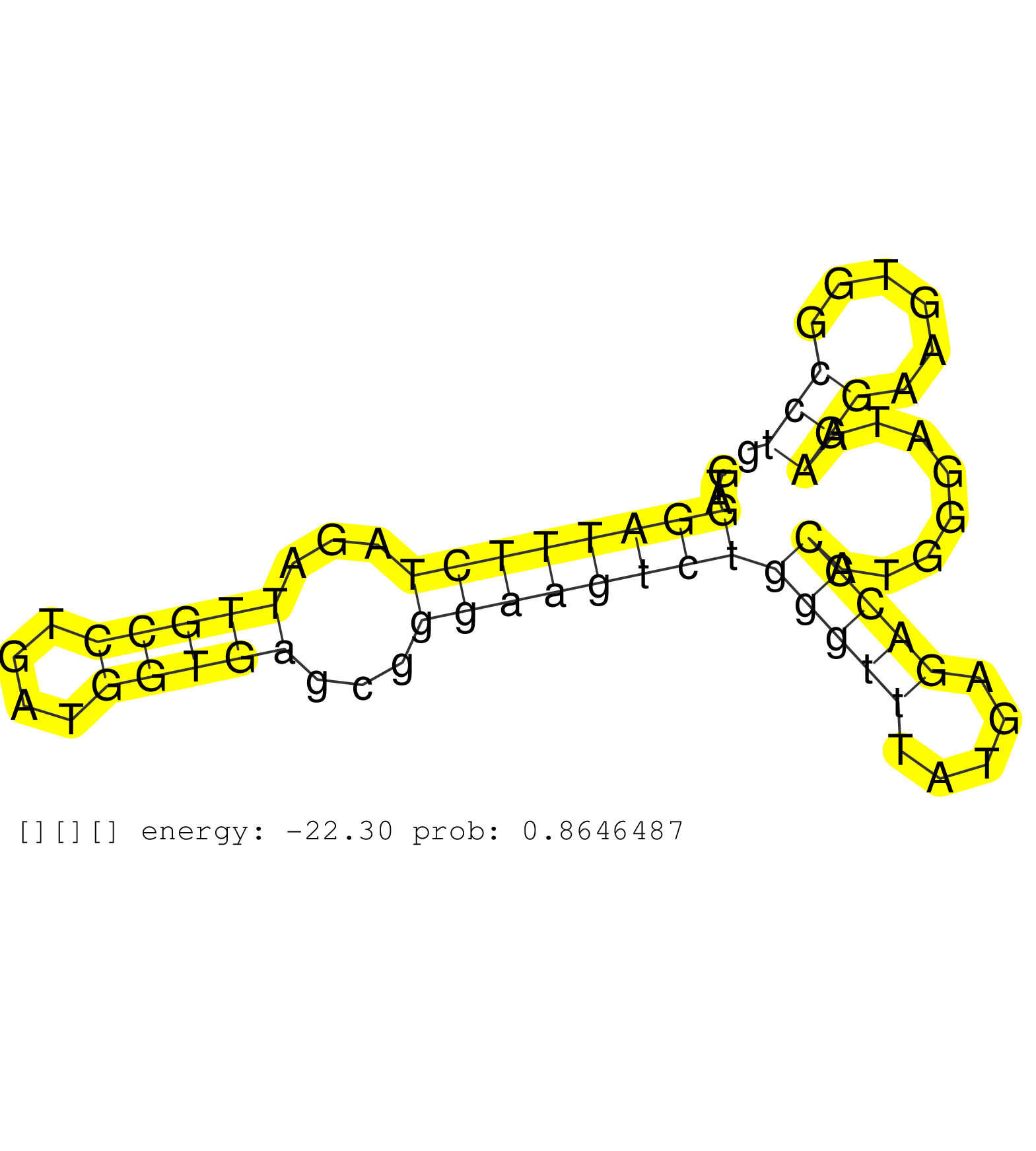

| Gene: Rage | ID: uc007pbv.1_intron_2_0_chr12_112048234_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CGAATTGCGGCTCACCAAGCCCTTCAGCACCCCTATTTCCAGGTGCAGAGGTAGGATTTCTAGATTGCCTGATGGTGAGCGGGAAGTCTGGGTTTATGAGACCCATGGGATAAGGAAGTGGCCTGGGAACGCTCTGTCCTGACAGTGATCATAGCCTTTGGGACCTTCTCTGTGATGTGGCCACCTCCACTAATGACCTTCCCAAATGAGGCAGTGAGCAGCTTCCTGCCCAGGGATAAAGGCTCAATGG .....................................................((((((((...(((((....)))))...))))))))(((((.....)))))........(((......))).............................................................................................................................. ..................................................51........................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGATTTCTAGATTGCCTGATGGTG............................................................................................................................................................................. | 27 | 1 | 10.00 | 10.00 | 2.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - |
| ..................................................GTAGGATTTCTAGATTGCCTGATGGT.............................................................................................................................................................................. | 26 | 1 | 7.00 | 7.00 | 3.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGATTTCTAGATTGCCTGATGGTGt............................................................................................................................................................................ | 28 | t | 3.00 | 10.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGATTTCTAGATTGCCTGATGGTGA............................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TATGAGACCCATGGGATAAGGAAGTGG................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TGAGACCCATGGGATAAGGAAGTGGCC............................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGGATAAGGAAGTGGCCTGGGAACGC...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGGATAAGGAAGTGGCCTGGGAAC........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGATTTCTAGATTGCCTGATGGTG............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGATTTCTAGATTGCCTGATGGTGAG........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................AAGGAAGTGGCCTGGGAACGCTCTGTCC............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................TTTCCAGGTGCAGAGGTAGGATTTCTAG........................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAGGATTTCTAGATTGCCTGATGGTGA............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGACAGTGATCATAGCCTTTGGGACCTT................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CGAATTGCGGCTCACCAAGCCCTTCAGCACCCCTATTTCCAGGTGCAGAGGTAGGATTTCTAGATTGCCTGATGGTGAGCGGGAAGTCTGGGTTTATGAGACCCATGGGATAAGGAAGTGGCCTGGGAACGCTCTGTCCTGACAGTGATCATAGCCTTTGGGACCTTCTCTGTGATGTGGCCACCTCCACTAATGACCTTCCCAAATGAGGCAGTGAGCAGCTTCCTGCCCAGGGATAAAGGCTCAATGG .....................................................((((((((...(((((....)))))...))))))))(((((.....)))))........(((......))).............................................................................................................................. ..................................................51........................................................................125........................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................CTTCCCAAATGAGGCAGt.................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |