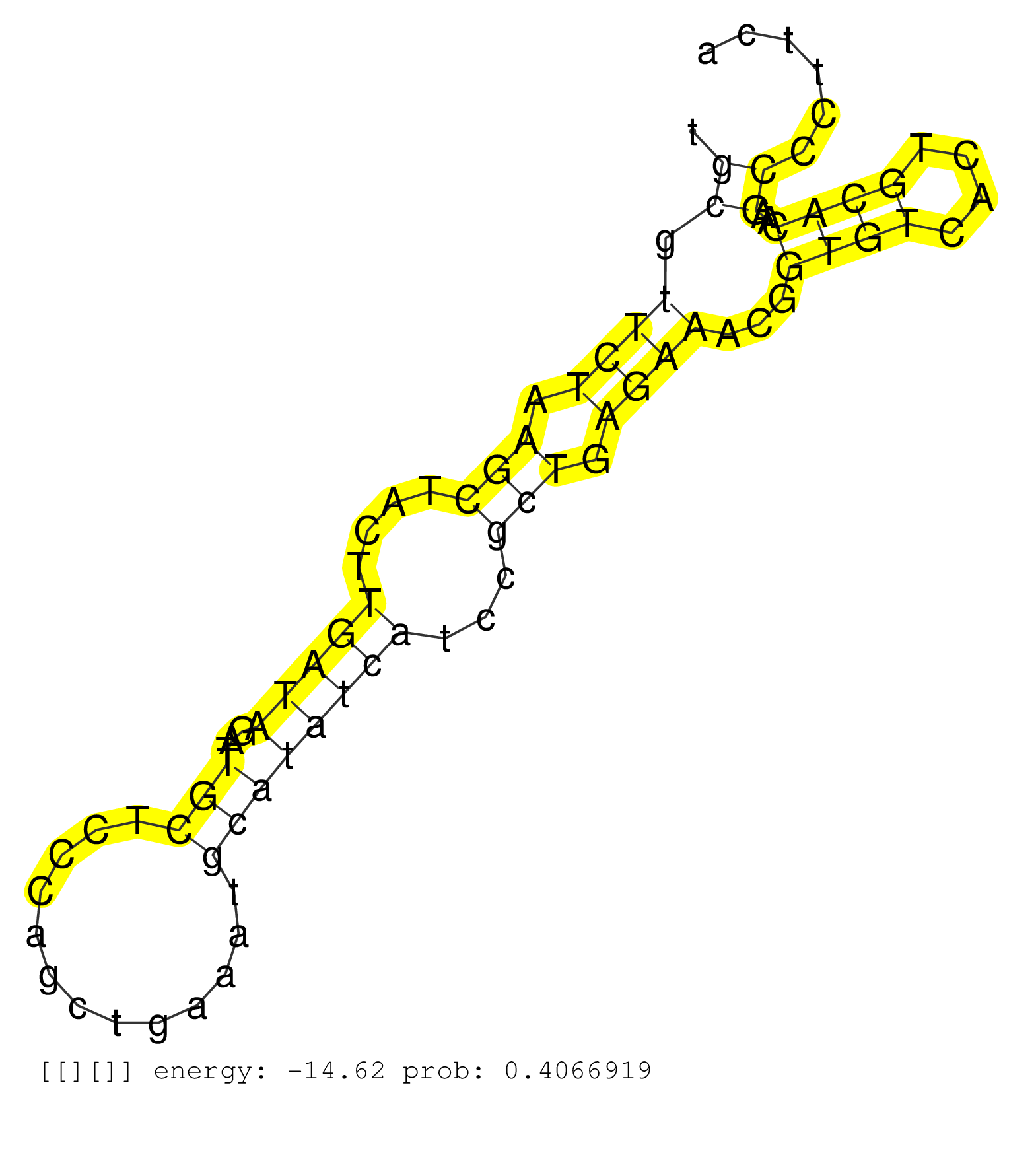

| Gene: 4933433P14Rik | ID: uc007oys.1_intron_1_0_chr12_106937187_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| CTTAAAGAGCAATGGGGGAAGCAAGGCACATTGTGGGTAGTGGTCCTGCGTTCTAAGCTACTTGATAGATTGCTCCCAGCTGAAATGCATATCATCCGCTGAGAAACGGTGTCACTGCACAAGCCCTTCAGGCAGCCGACCCTTGCCTTGTGTGCCAGGATGGACGCTGACCACTGCTTACTGCTGCCCTTCTCTCGTAGGTGGTGGGCTATGCTTTTGACCAGGTGGAGGATCATTTGCAAACCCCCTA ...............................................((.((((.(((....(((((...(((.............))))))))...))).))))...((((....))))..)).............................................................................................................................. ..............................................47.................................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TGAGAAACGGTGTCACTGCACAAGCCC............................................................................................................................ | 27 | 1 | 9.00 | 9.00 | 4.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................TGATAGATTGCTCCCAGCTGAAATGCATA............................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................TGATAGATTGCTCCCAGCTGAAATGCAT................................................................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TCTAAGCTACTTGATAGATTGCTCCC............................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGGGCTATGCTTTTGACCAGGTGGAGG................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGCTTTTGACCAGGTGGAGGATCATT............. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ACCAGGTGGAGGATCctgt............ | 19 | ctgt | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| .......................................................................................................................................................................TGACCACTGCTTACTGCTGCCCTTCTC........................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGAGAAACGGTGTCACTGCACAAGCCCT........................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGGACGCTGACCACTGCTTACTGCTGCC.............................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGCTTTTGACCAGGTGGAGGATCAT.............. | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGGGCTATGCTTTTGACCAGGTGGAG.................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TCCGCTGAGAAACGGTGTCACTGCACAAG............................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGGGTAGTGGTCCTGCGTTCTAAGCT............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGACCAGGTGGAGGATCATTTGCAAACCCCCT. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................TGAGAAACGGTGTCACTGCACAAGCtcc........................................................................................................................... | 28 | tcc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TAGATTGCTCCCAGCTGAAATGCATATC............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................TTGCCTTGTGTGCCAGGATGGACGCTG................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTCTCTCGTAGGTGGTGGGCTATGC.................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................TACTTGATAGATTGCTCCCAGCTGA....................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................TGCTTTTGACCAGGTGGAGGATCATTTG........... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TATCATCCGCTGAGAAACGGTGTCACTGC.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AAGAGCAATGGGGGAAGCAAGG................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................AAACGGTGTCACTGCACAAGCCCTTCA........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTAAAGAGCAATGGGGGAAGCAAGGC............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................TACTTGATAGATTGCTCCCAGCTGAAATGCAT................................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..TAAAGAGCAATGGGGGAAGCAAGGCA.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..TAAAGAGCAATGGGGGAAGCAAGGC............................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAAATGCATATCATCCGCTGAGAAAC............................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................TAGTGGTCCTGCGTTCTAAGCTACTT........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ACCAGGTGGAGGATCctgc............ | 19 | ctgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................GGCACATTGTGGGTAGTGGTCCTGC......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................CTGCGTTCTAAGCTACTTGATAGATTGC................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTAAAGAGCAATGGGGGAAGCAAGGCACATTGTGGGTAGTGGTCCTGCGTTCTAAGCTACTTGATAGATTGCTCCCAGCTGAAATGCATATCATCCGCTGAGAAACGGTGTCACTGCACAAGCCCTTCAGGCAGCCGACCCTTGCCTTGTGTGCCAGGATGGACGCTGACCACTGCTTACTGCTGCCCTTCTCTCGTAGGTGGTGGGCTATGCTTTTGACCAGGTGGAGGATCATTTGCAAACCCCCTA ...............................................((.((((.(((....(((((...(((.............))))))))...))).))))...((((....))))..)).............................................................................................................................. ..............................................47.................................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|