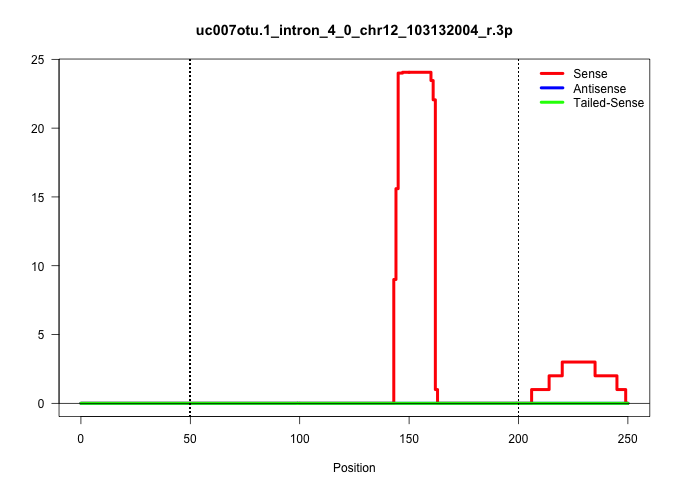
| Gene: AK081763 | ID: uc007otu.1_intron_4_0_chr12_103132004_r.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) PIWI.ip |
(14) TESTES |
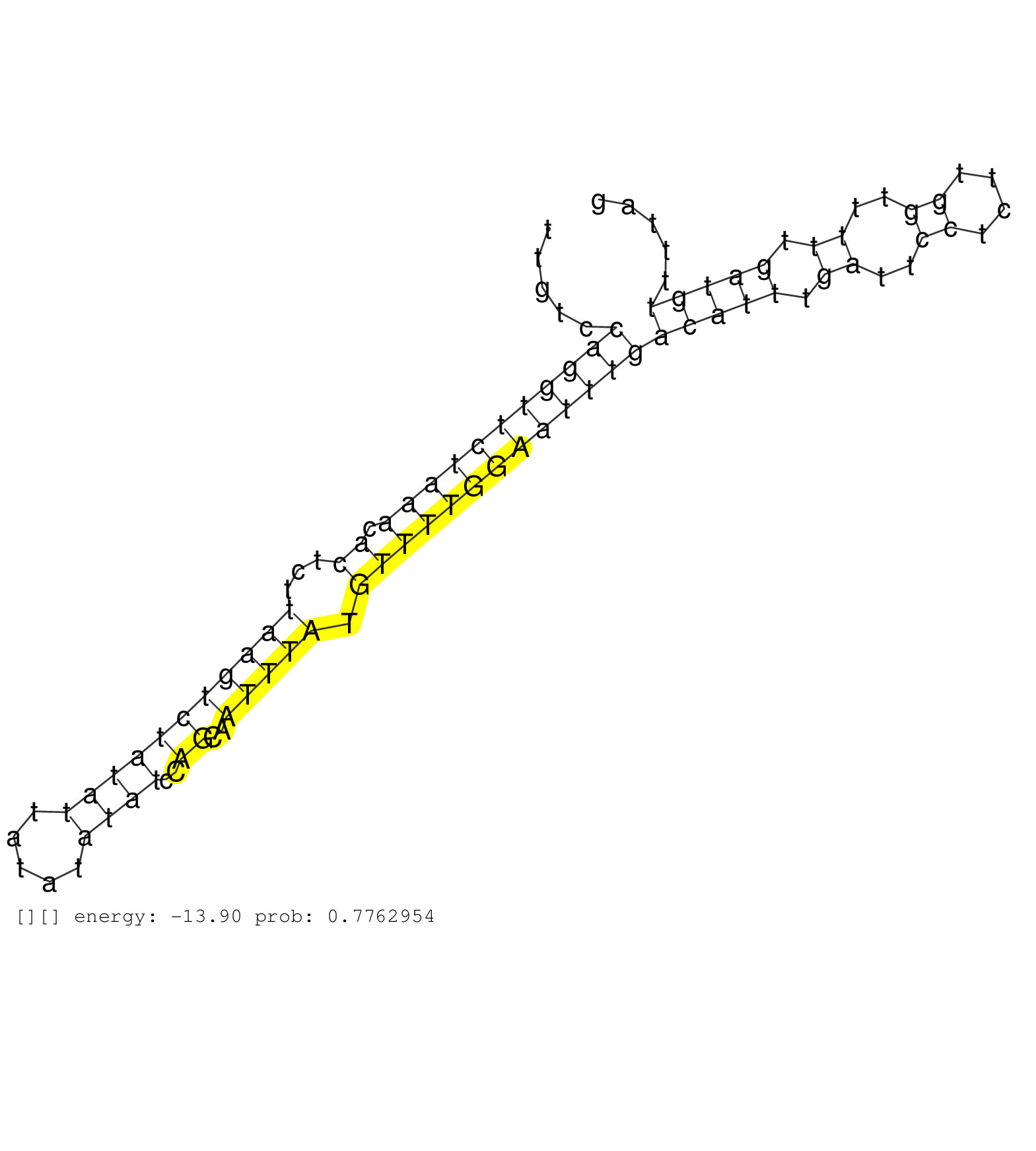
| ACCAAGATATTTTTACCTTGTTTTAAACTCAAATATATAGACTTAATAGTTCAAATGAAGTCTTTGACTTTTTATTTATTTAAAATTCTAGTAATTTAAGTTGTCCAGGTTCTAAACACTCTTAAGTCTATATTATATATATCCAGCAATTTATGTTTTGGAATTTGACATTTGATTCCTCTTGGTTTTTGATGTTTTAGGTGGCTCAGGAGTTACGGTAACTGATCACTCTAAAGTGTATGAGATGCAA .........................................................................................................(((((((((((.((...(((((((((((.....))))..))..))))).)))))))))))))(((((.((..((....))..)).)))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................GCAATTTATGTTTTGGA........................................................................................ | 17 | 1 | 11.00 | 11.00 | 8.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................CAGCAATTTATGTTTTGGA........................................................................................ | 19 | 1 | 8.00 | 8.00 | 3.00 | 1.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................AGCAATTTATGTTTTGGA........................................................................................ | 18 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGCAATTTATGTTTTGGAA....................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................ACTGATCACTCTAAAGTGTATGAGATGCA. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTCCAGGTTCTAAACACTCTTAAGTCTAT....................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................CAGCAATTTATGTTTTGG......................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................ACGGTAACTGATCACTCTAAAGTGTATGAGA..... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................CAGGAGTTACGGTAACTGATCACTCTAAA............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................AGCAATTTATGTTTTG.......................................................................................... | 16 | 5 | 0.60 | 0.60 | 0.60 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GCAATTTATGTTTTGG......................................................................................... | 16 | 5 | 0.40 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................AATTTATGTTTTGGA........................................................................................ | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 |
| ACCAAGATATTTTTACCTTGTTTTAAACTCAAATATATAGACTTAATAGTTCAAATGAAGTCTTTGACTTTTTATTTATTTAAAATTCTAGTAATTTAAGTTGTCCAGGTTCTAAACACTCTTAAGTCTATATTATATATATCCAGCAATTTATGTTTTGGAATTTGACATTTGATTCCTCTTGGTTTTTGATGTTTTAGGTGGCTCAGGAGTTACGGTAACTGATCACTCTAAAGTGTATGAGATGCAA .........................................................................................................(((((((((((.((...(((((((((((.....))))..))..))))).)))))))))))))(((((.((..((....))..)).)))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................TTAATAGTTCAAATGaaa................................................................................................................................................................................................. | 18 | aaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TTCTAAACACTCTTAAggac............................................................................................................................. | 20 | ggac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................TTTATGTTTTGGAAggtc....................................................................................... | 18 | ggtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |