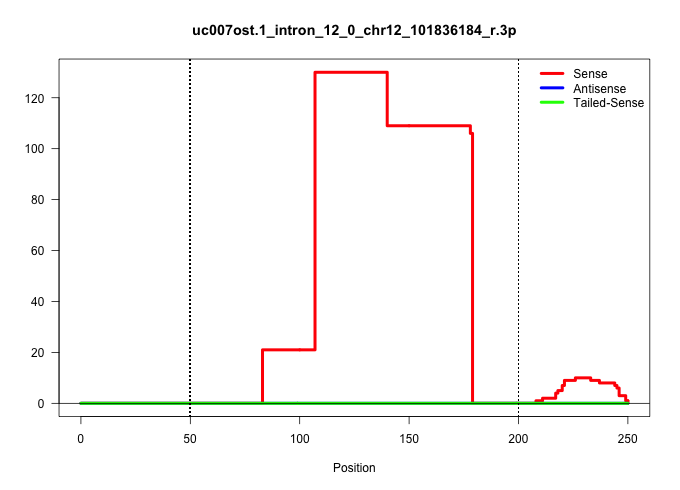
| Gene: Rps6ka5 | ID: uc007ost.1_intron_12_0_chr12_101836184_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(13) TESTES |
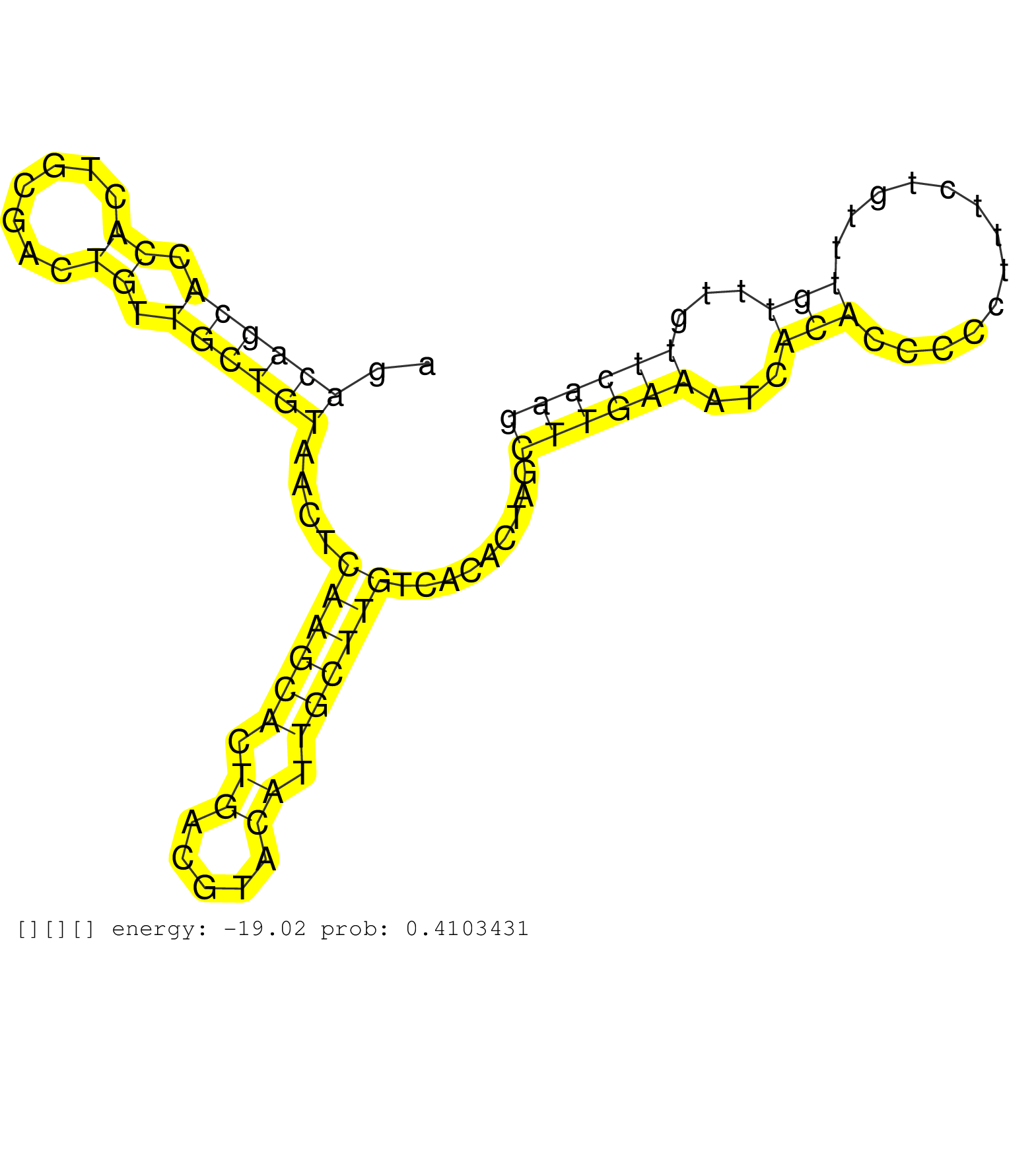
| GGTTGATGGATGCTAGCCATCAGTGTTTCAGGCAAAATGTTTACCCAGTGCCCTCCATAGCAGGCAGAGCCTGTTCTGTCCTGATAGAGGTCTGTCGGGCAGACAGCACCACTGCGACTGTTGCTGTAACTCAAGCACTGACGTACATTGCTTGTCACACTAGCTTGAAATCACACCCCCTTTCTGTTTGTTTGTTCAAGACTGAAAGAGCATATTCTTTTTGTGGAACTATTGAATACATGGCTCCAGA ......................................................................................................((((((.((.......)).))))))....((((((.((.....)).)))))).........((((((...(((.............)))...)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................ACCACTGCGACTGTTGCTGTAACTCAAGCACTGACGTACATTGCTTGTCACA........................................................................................... | 52 | 1 | 109.00 | 109.00 | 109.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................ATAGAGGTCTGTCGGGCAGACAGCACCACTGCGACTGTTGCTGTAACTCAAG................................................................................................................... | 52 | 1 | 21.00 | 21.00 | - | 21.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTGTGGAACTATTGAATACATGGCTC.... | 26 | 1 | 5.00 | 5.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................................TGTGGAACTATTGAATACATGGCTCCAG. | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................TATTCTTTTTGTGGAACTATTGAAT............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................AACTATTGAATACATGGCTCCAGA | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTTTTGTGGAACTATTGAATACATGGC...... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGCATATTCTTTTTGTGGAACTATT................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTGTTTGTTTGTTCAAGA................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TTTTTGTGGAACTATTGAATACATGGCT..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGAAAGAGCATATTCTTTTTGTGGAA...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................ATATTCTTTTTGTGGAACTATTGAAT............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TTTTGTGGAACTATTGAATACATGGCTC.... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GGTTGATGGATGCTAGCCATCAGTGTTTCAGGCAAAATGTTTACCCAGTGCCCTCCATAGCAGGCAGAGCCTGTTCTGTCCTGATAGAGGTCTGTCGGGCAGACAGCACCACTGCGACTGTTGCTGTAACTCAAGCACTGACGTACATTGCTTGTCACACTAGCTTGAAATCACACCCCCTTTCTGTTTGTTTGTTCAAGACTGAAAGAGCATATTCTTTTTGTGGAACTATTGAATACATGGCTCCAGA ......................................................................................................((((((.((.......)).))))))....((((((.((.....)).)))))).........((((((...(((.............)))...)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................GGGCAGACAGCACCActtt........................................................................................................................................... | 19 | cttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |