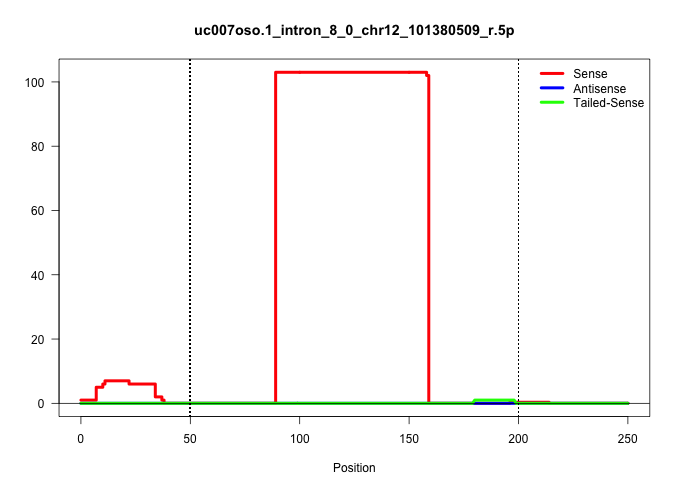
| Gene: BC002230 | ID: uc007oso.1_intron_8_0_chr12_101380509_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
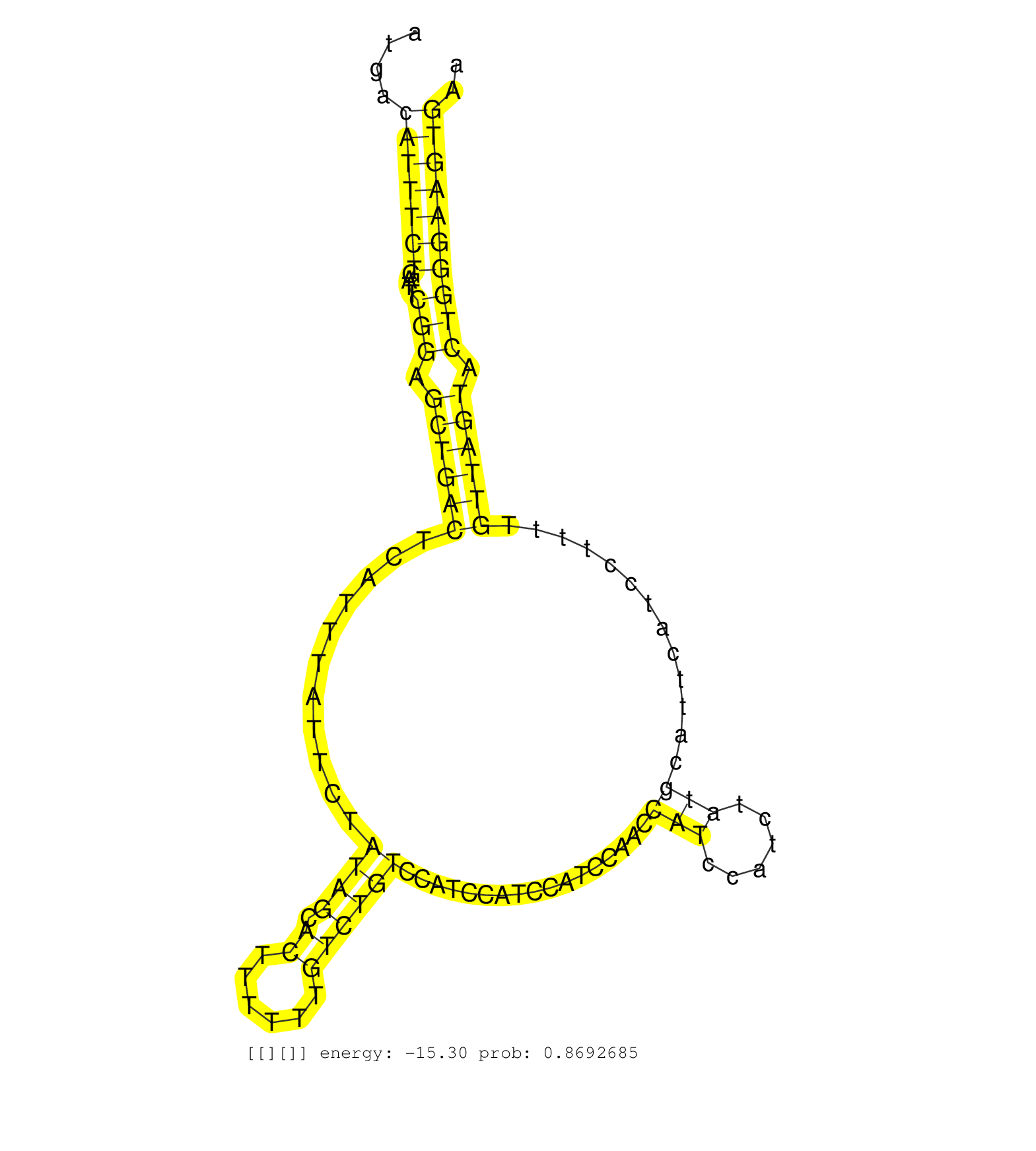
| GGGAGAATCCTTGGGACACGCAACTATGGATGGCGTTTGTGGCTTTCCAGGTAAGTGTCACTGTCCTGGACCTCTTACTTTGAAATGACATTTCTGATTCGGAGCTGACTCATTTATTCTATAGCACTTTTTTGTCTGTCCATCCATCCATCCAACCATCCATCTATGCATTCATCCTTTTGTTAGTACTGGGAAGTGAACCCAGGATCTCGAATATACTTGCATATATCAGTGTCTGCTACTGATCCCC ........................................................................................(((((((....(((.((((((...........((((.((......)))))).................(((......))).............)))))).)))))))))).................................................... ....................................................................................85.................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................ATTTCTGATTCGGAGCTGACTCATTTATTCTATAGCACTTTTTTGTCTGTCC............................................................................................................. | 52 | 1 | 103.00 | 103.00 | 103.00 | - | - | - | - | - | - | - | - | - |
| .......TCCTTGGGACACGCAACTATGGATGGC........................................................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTTAGTACTGGGAAcaca................................................... | 19 | caca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........TGGGACACGCAACTATGGATGGCGTTT.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .GGAGAATCCTTGGGACACGCA.................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........TTGGGACACGCAACTATGGATGGCGTT..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................TGAACCCAGGATCTCGAA.................................... | 18 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 |
| GGGAGAATCCTTGGGACACGCAACTATGGATGGCGTTTGTGGCTTTCCAGGTAAGTGTCACTGTCCTGGACCTCTTACTTTGAAATGACATTTCTGATTCGGAGCTGACTCATTTATTCTATAGCACTTTTTTGTCTGTCCATCCATCCATCCAACCATCCATCTATGCATTCATCCTTTTGTTAGTACTGGGAAGTGAACCCAGGATCTCGAATATACTTGCATATATCAGTGTCTGCTACTGATCCCC ........................................................................................(((((((....(((.((((((...........((((.((......)))))).................(((......))).............)))))).)))))))))).................................................... ....................................................................................85.................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CCATCCATCCATCCAACCATCCATCtat...................................................................................... | 28 | tat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................CCATCCATCCATCCAACCATCCATCca...................................................................................... | 27 | ca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................CCATCCATCCATCCAACCATtatt........................................................................................... | 24 | tatt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - |