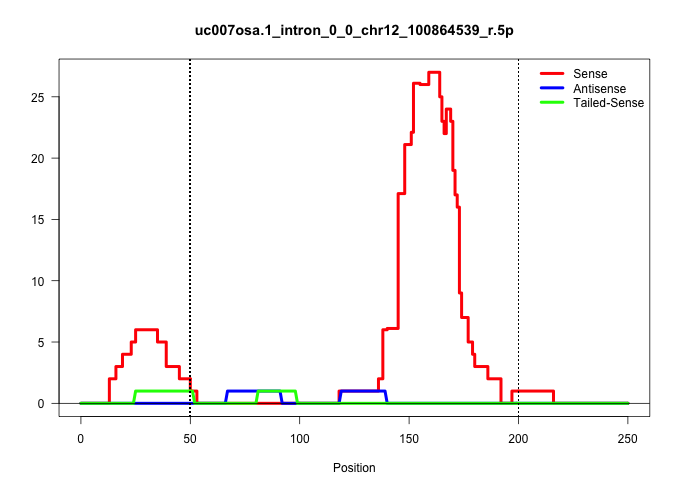
| Gene: AK006884 | ID: uc007osa.1_intron_0_0_chr12_100864539_r.5p | SPECIES: mm9 |
 |
|
(6) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TGAAAGACTTTTTTTTTTTTAAGTCTAAGAAAGAGTATATGCAGAAAACGGTGAGGCTTCCAAATTCTGTCTTCGAAAACAACGTTGAAATAAATGATCCATTTCTTAAGGGCCATTTTAAAATCGAAAATGTCCGTATACTGGATTCTGAATCCGGACTGGCAATCTGCAGTGGGCTTCACCCGGAGAATCCGGTTAGAGGTTTTAAAATTTAAACCGTTTACACCACCCCACTCCGCCCCTTGCTAGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TTCTGAATCCGGACTGGCAATCTGCAGT............................................................................. | 28 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCTGAATCCGGACTGGCAATCTGC................................................................................ | 25 | 1 | 4.00 | 4.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................TCCGGACTGGCAATCTGCAGTGGGC......................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGAATCCGGACTGGCAATCTGCAGTG............................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGCAGTGGGCTTCACCCGGAGAATC.......................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TTTTTTTAAGTCTAAGAAAGAGTATA................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TACTGGATTCTGAATCCGGACTGGCAA..................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................TCCGGACTGGCAATCTGCAGTGGGCTTC...................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TAAGAAAGAGTATATGCAGAAAACGGg...................................................................................................................................................................................................... | 27 | g | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TACTGGATTCTGAATCCGGACTGGCAAT.................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TATACTGGATTCTGAATCCGGACTGGCA...................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................TAAAATCGAAAATGTCCGTATACTGGA......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGGCAATCTGCAGTGGGCTTCACCCGG................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................TCCGGACTGGCAATCTGCAGTGGGCTT....................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCTGAATCCGGACTGGCAATCTGCAG.............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................TTTTAAGTCTAAGAAAGAG....................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGAATCCGGACTGGCAATCTGCAGT............................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................ACGTTGAAATAAATctct....................................................................................................................................................... | 18 | ctct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................TAAGTCTAAGAAAGAGTATATGCAGA............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................AGAGGTTTTAAAATTTAAA.................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................ATCCGGACTGGCAATCTG................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................TACTGGATTCTGAATCCGGACTGGCA...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTAAGAAAGAGTATATGCAGAAAACG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCTGAATCCGGACTGGCAATCTGCA............................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TAAGAAAGAGTATATGCAGAAAACGGTG..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGAATCCGGACTGGCAATCTGCA............................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGATTCTGAATCC............................................................................................... | 15 | 19 | 0.11 | 0.11 | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 |
| TGAAAGACTTTTTTTTTTTTAAGTCTAAGAAAGAGTATATGCAGAAAACGGTGAGGCTTCCAAATTCTGTCTTCGAAAACAACGTTGAAATAAATGATCCATTTCTTAAGGGCCATTTTAAAATCGAAAATGTCCGTATACTGGATTCTGAATCCGGACTGGCAATCTGCAGTGGGCTTCACCCGGAGAATCCGGTTAGAGGTTTTAAAATTTAAACCGTTTACACCACCCCACTCCGCCCCTTGCTAGG |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................AAAATCGAAAATGTCCGTATA.............................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................TGTCTTCGAAAACAACGTTGAAATA.............................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |