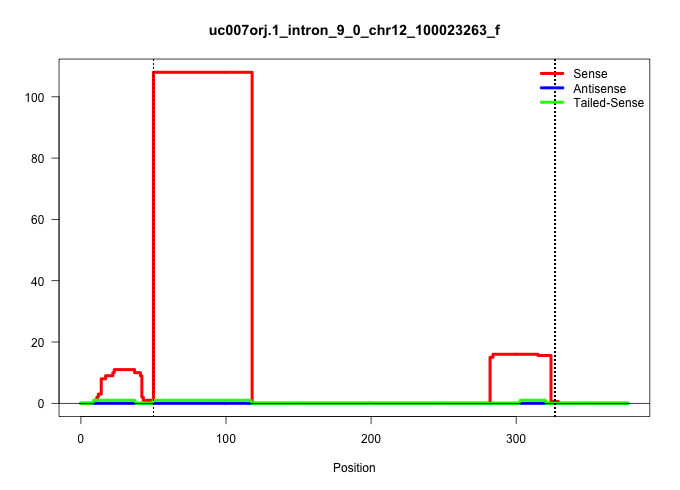
| Gene: Zc3h14 | ID: uc007orj.1_intron_9_0_chr12_100023263_f | SPECIES: mm9 |
 |
 |
|
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
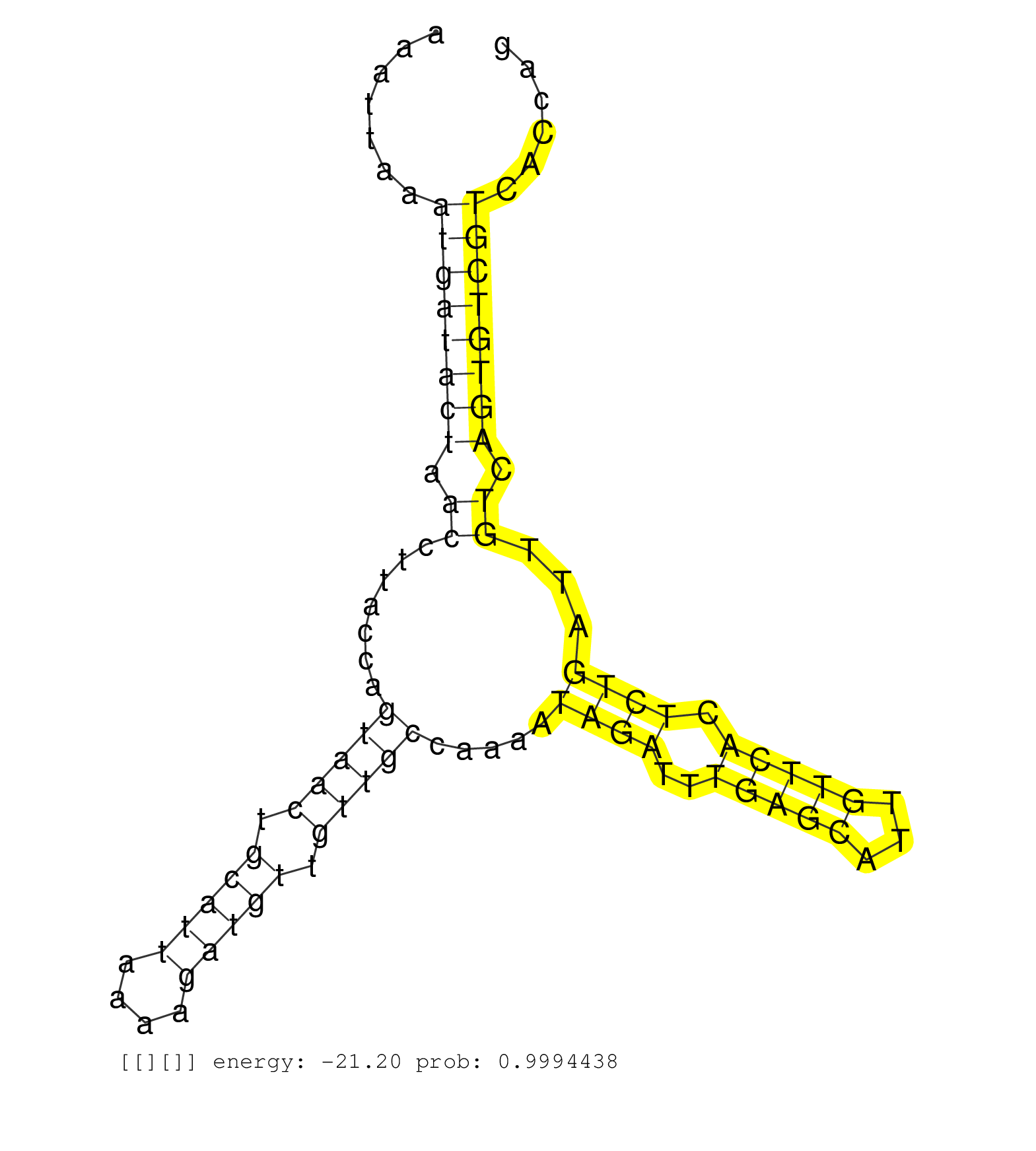
| TCCCTTCACTCACATGAGTAGAAGAGCCTCGATACTGACTCCAAAACCAGGTTAGTAACTTTGTGCAGTTATAGTCAGGTCAAAAAGAAGAAAACGACATTTTCTGGCTCTAAGTGATAGGAAACAACAAAAGAATAGTTTTAGAAATGATTTTAGTATTTAAAGAACAAATCTGCCTTTCTTACAGAACTCTGCTATATTTTATCTTGACTGATAAAACTTGTTCTAAATTAAATGATACTAACCTTACCAGTAACTGCATTAAAAGATGTTGTTGCCAAAATAGATTTGAGCATTGTTCACTCTGATTGTCAGTGTCGTCACCAGCACCGTCTTCTAATGGCCAGCTCTGCCGTTACTTCCCTGCTTGTAAGAAA ..........................................................................................................................................................................................................................................((((((((.((.......(((((.(((((....))))).))))).....((((..(((((...))))).))))...)).))))))))........................................................ ...................................................................................................................................................................................................................................228................................................................................................327................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTTAGTAACTTTGTGCAGTTATAGTCAGGTCAAAAAGAAGAAAACGACATTT................................................................................................................................................................................................................................................................................... | 52 | 1 | 109.00 | 109.00 | 109.00 | - | - | - | - | - | - | - | - | - | - |
| ..............TGAGTAGAAGAGCCTCGATACTGACTCC............................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - |
| ...........ACATGAGTAGAAGAGCCTCGATACTGACTCCA.............................................................................................................................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............CATGAGTAGAAGAGCCTCGATACTGACTC................................................................................................................................................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................GTAGAAGAGCCTCGATACTGACTCC............................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........TCACATGAGTAGAAGAGCCTCGATACTGt................................................................................................................................................................................................................................................................................................................................................... | 29 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................GAGCCTCGATACTGACTCC............................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................TCTGATTGTCAGTGaggt........................................................ | 18 | aggt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................AGATTTGAGCATTGTTCACTCTGATTGTCAG.............................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................AGAGCCTCGATACTGACTCCAAAACCAG....................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................CAACAAAAGAATAGTTTTAGAAATGAT.................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........TCACATGAGTAGAAGAGCCTCGATACTG.................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................TCACTCTGATTGTCAG.............................................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | 0.25 |
| TCCCTTCACTCACATGAGTAGAAGAGCCTCGATACTGACTCCAAAACCAGGTTAGTAACTTTGTGCAGTTATAGTCAGGTCAAAAAGAAGAAAACGACATTTTCTGGCTCTAAGTGATAGGAAACAACAAAAGAATAGTTTTAGAAATGATTTTAGTATTTAAAGAACAAATCTGCCTTTCTTACAGAACTCTGCTATATTTTATCTTGACTGATAAAACTTGTTCTAAATTAAATGATACTAACCTTACCAGTAACTGCATTAAAAGATGTTGTTGCCAAAATAGATTTGAGCATTGTTCACTCTGATTGTCAGTGTCGTCACCAGCACCGTCTTCTAATGGCCAGCTCTGCCGTTACTTCCCTGCTTGTAAGAAA ..........................................................................................................................................................................................................................................((((((((.((.......(((((.(((((....))))).))))).....((((..(((((...))))).))))...)).))))))))........................................................ ...................................................................................................................................................................................................................................228................................................................................................327................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................CAGAACTCTGCTATAatg.................................................................................................................................................................................. | 18 | atg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |