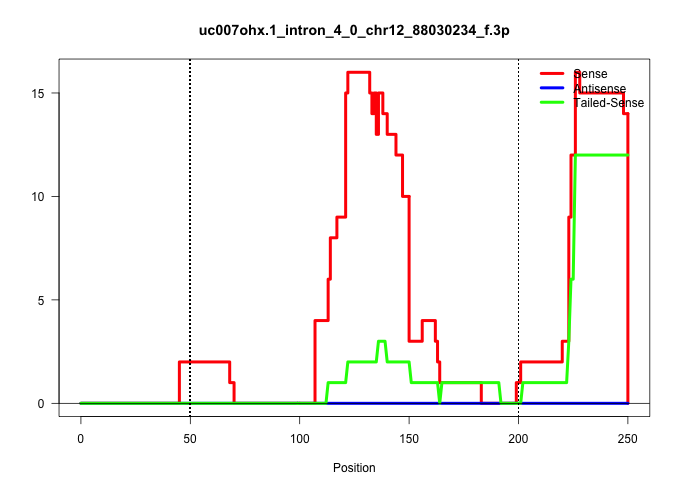
| Gene: Vash1 | ID: uc007ohx.1_intron_4_0_chr12_88030234_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| CCTACCAAGGATCTGCTGCCCGCCTCCCCCCCCCCCCCATGGCTTTAGTGCCAGGGATGACCCAGAGATGGGGTAAAGGCCACCTGAGGCTTGGAGCTCAGACCGCATAGGGATGGCTTCCAGGGACTGTGACTCCTAGCAGGACAGAGTAGGCCATGCGGCAGCTCAGAACATCCTTTTCTCTCTTCCTCCCTATCCAGATTGGCAAGGGGACAGGCCCTCCTTCTCCCACCAAGGACCGGAAGAAGGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................AGGGACTGTGACTCCTAGCAGGACAGAGT.................................................................................................... | 29 | 1 | 6.00 | 6.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTCTCCCACCAAGGACCGGAAGAAGGA | 27 | 1 | 6.00 | 6.00 | - | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGA | 24 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCTCCCACCAAGGACCGGAAGAAGGA | 26 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTCTCCCACCAAGGACCGGAAGAAGGAtt | 29 | tt | 2.00 | 6.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGAtgaa | 28 | tgaa | 2.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCTCCCACCAAGGACCGGAAGAAGGAtt | 28 | tt | 2.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................TAGGGATGGCTTCCAGGGACTGTGACTC................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GGCTTCCAGGGACTGTGACTCCTAGCAGGACAG....................................................................................................... | 33 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .............................................TAGTGCCAGGGATGACCCAGAGATG.................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGAagt | 27 | agt | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCTCCCACCAAGGACCGGAAGAAGGAta | 28 | ta | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGCTTCCAGGGACTGTGACTCCTAGa.............................................................................................................. | 27 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TAGGGATGGCTTCCAGGGACTGTGA...................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGAta | 26 | ta | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCTAGCAGGACAGAGTAGGCCATGCGGC........................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGCTTCCAGGGACTGTGACTCCTAGC.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGGCAAGGGGACAGGCCCTCCTTCTC...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................TAGTGCCAGGGATGACCCAGAGA...................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGAcg | 26 | cg | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GGGACTGTGACTCCTAGCAGGACAGAGT.................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GGGACTGTGACTCCTAGCAGGACAGAGat................................................................................................... | 29 | at | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATTGGCAAGGGGACAGGCCCTCCT.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................TCCTTCTCCCACCAAGGACCGGAAGAAG.. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................TAGCAGGACAGAGTAGGCCATGCGGCA....................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGtatg | 26 | tatg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TCCCACCAAGGACCGGAAGAAGGAtga | 27 | tga | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCAGAACATCCTTTTCTCTCTTCCTCa.......................................................... | 27 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TAGCAGGACAGAGTAGGCCATGCGGCAG...................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGCTTCCAGGGACTGTGACTCCTA................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGGCAAGGGGACAGGCCCTCCTTa........................ | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................TGCGGCAGCTCAGAACATCCTTTTCTC................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TTCCAGGGACTGTGACTCCTAGCAGGA.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TAGGGATGGCTTCCAGGGACTGTGAC..................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TAGCAGGACAGAGTAGGCCATGCGGCcg...................................................................................... | 28 | cg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTACCAAGGATCTGCTGCCCGCCTCCCCCCCCCCCCCATGGCTTTAGTGCCAGGGATGACCCAGAGATGGGGTAAAGGCCACCTGAGGCTTGGAGCTCAGACCGCATAGGGATGGCTTCCAGGGACTGTGACTCCTAGCAGGACAGAGTAGGCCATGCGGCAGCTCAGAACATCCTTTTCTCTCTTCCTCCCTATCCAGATTGGCAAGGGGACAGGCCCTCCTTCTCCCACCAAGGACCGGAAGAAGGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................CCATGCGGCAGCTCAgct.................................................................................. | 18 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |