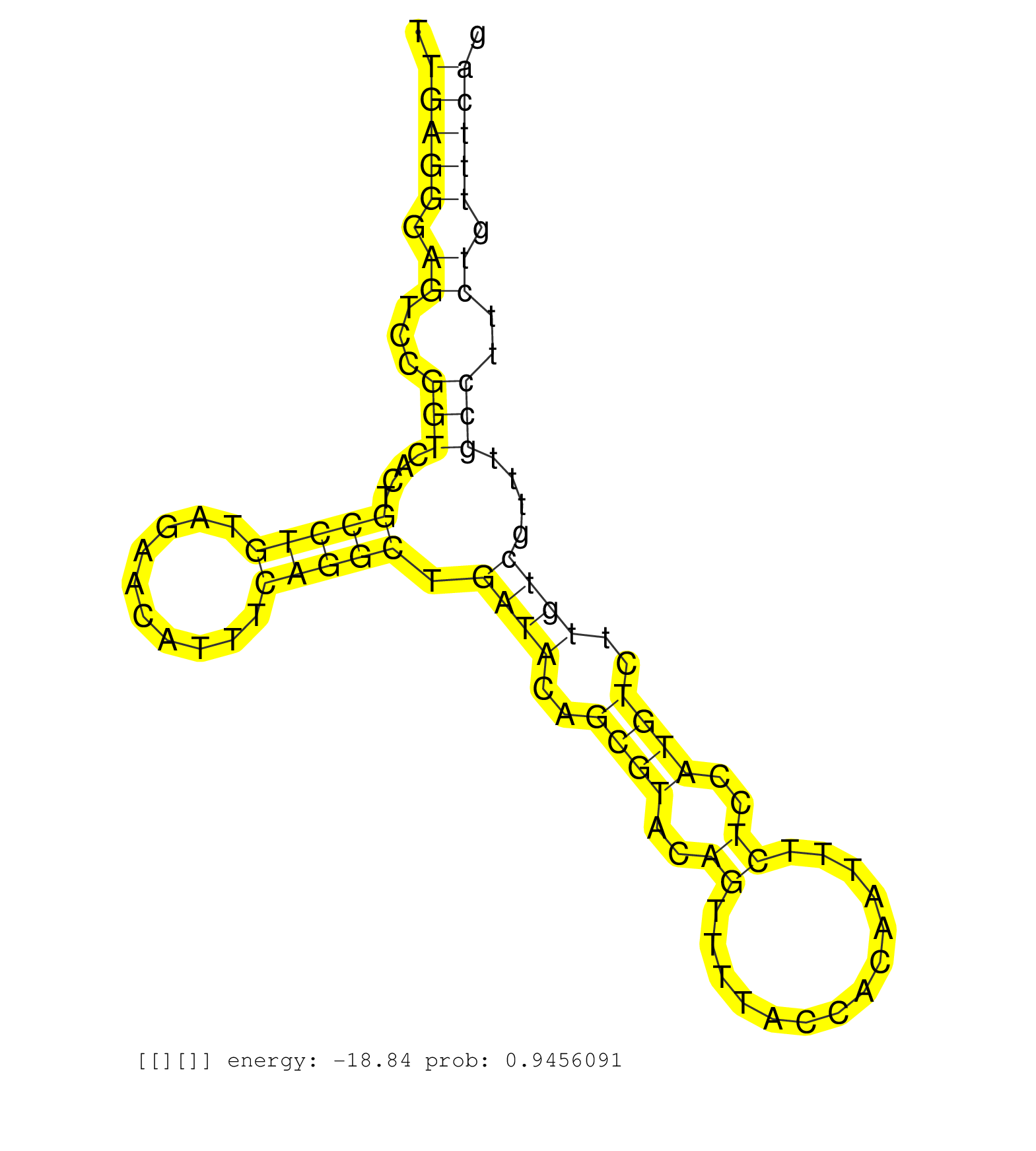

| Gene: 1110018G07Rik | ID: uc007ofy.1_intron_4_0_chr12_86265076_r.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| AGTCTTAGCTAGTGGAGTGTGAATAGTGGTGTGAGCCCTGAAAGTCAGTAACAGAGAGTGAAAAGCAACTGTGTTAAAAAGGCTAATGCAGATGAGAAACTTGAGGGAGTCCGGTCACTGCCTGTAGAACATTTCAGGCTGATACAGCGTACAGTTTTACCACAATTTCTCCATGTCTTGTCGTTTGCCTTCTGTTTCAGATTGTAGAACTCATGACAGGCGGAGCTCAAACCCCAGTCACCAATGCAAA .....................................................................................................(((((.((...(((....(((((..........))))).((((..((((..((..............))..))))..))))....)))..)).)))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................AGATGAGAAACTTGAGGGAGTCCGGTCACTGCCTGTAGAACATTTCAGGCTG............................................................................................................. | 52 | 1 | 105.00 | 105.00 | 105.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGTAGAACTCATGACAGGCGGA.......................... | 23 | 1 | 7.00 | 7.00 | - | - | - | 7.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGCTC...................... | 26 | 1 | 6.00 | 6.00 | - | 2.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TAGAACTCATGACAGGCGGAGCTCAA.................... | 26 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGTAGAACTCATGACAGGCGGAGCT....................... | 26 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGCTCA..................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................TGACAGGCGGAGCTCAAACCCCAG............. | 24 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TAGAACTCATGACAGGCGGAGCTCAAACCC................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGACAGGCGGAGCTCAAACC................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................TTTCAGATTGTAGAACTCATGACAGGCG............................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGATTGTAGAACTCATGACAGGCGGA.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................AGATTGTAGAACTCATGACAGGCGGAG......................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TAGAACTCATGACAGGCGGAGCTCAAAC.................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGCTCt..................... | 27 | t | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGATTGTAGAACTCATGACAGGCGGAGC........................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GAACTCATGACAGGCGGAGCTCAAACCC................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATTGTAGAACTCATGACAGGCGGAGC........................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGCTCAAt................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGATTGTAGAACTCATGACAGGCGGggt........................ | 28 | ggt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGgtc...................... | 26 | gtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TAGAACTCATGACAGGCGGAGCTCAAA................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTAGAACTCATGACAGGCGGAGC........................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| AGTCTTAGCTAGTGGAGTGTGAATAGTGGTGTGAGCCCTGAAAGTCAGTAACAGAGAGTGAAAAGCAACTGTGTTAAAAAGGCTAATGCAGATGAGAAACTTGAGGGAGTCCGGTCACTGCCTGTAGAACATTTCAGGCTGATACAGCGTACAGTTTTACCACAATTTCTCCATGTCTTGTCGTTTGCCTTCTGTTTCAGATTGTAGAACTCATGACAGGCGGAGCTCAAACCCCAGTCACCAATGCAAA .....................................................................................................(((((.((...(((....(((((..........))))).((((..((((..((..............))..))))..))))....)))..)).)))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................GATTGTAGAACTCcga...................................... | 16 | cga | 3.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................AAACCCCAGTCACCAATGct.... | 20 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |