
| Gene: 2900006K08Rik | ID: uc007ofi.1_intron_10_0_chr12_85771284_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) PIWI.ip |
(11) TESTES |
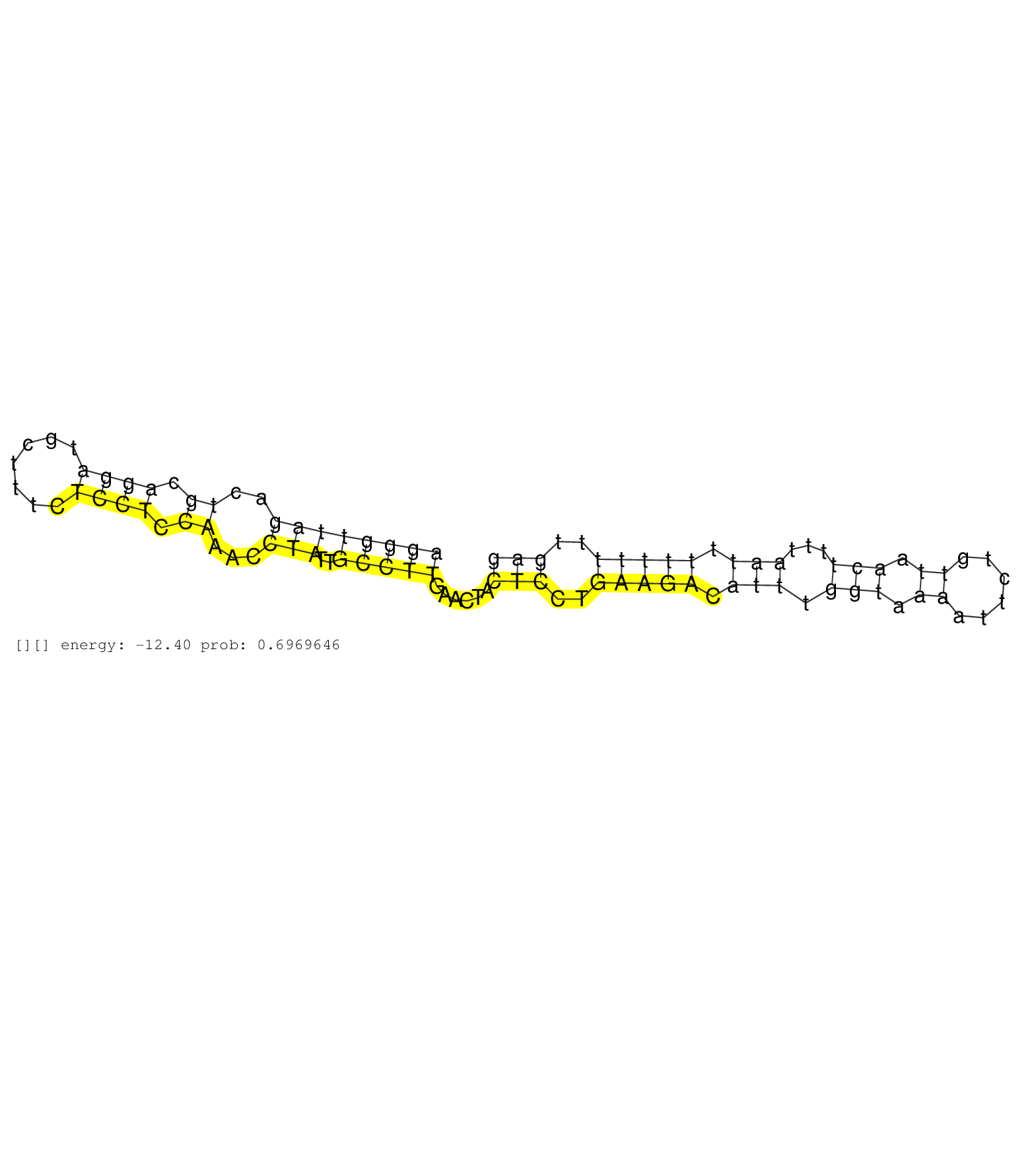
| TCCGAAAAAAAAAATAATCAATCTTCGGATACGTGTTTGTGGGTAAAGCAGTTGCCACATGACCATTAGGACCTGGCTCCGACTCCCAGAACCCATGGAAAGGGTTAGACTGCAGGATGCTTTCTCCTCCAAACCTATTGCCTTGAACTACTCCTGAAGACATTTGGTAAAATTCTGTTAACTTTTAATTTTTTTTTGAGGCCTAGGAAGTAGAGCAGCACAGCAGATGACTCCACAGTAGATGCAGGAA ....................................................................................................((((((((..((.((((.......)))).))...)))..)))))......(((..(((((.(((.(((.((......)).)))...))).)))))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................CTCCTCCAAACCTATTGCCTTGAACTACTCCTGAAGACATTTGGTAAAATTC........................................................................... | 52 | 1 | 28.00 | 28.00 | 28.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GAAGTAGAGCAGCACAGCAG........................ | 20 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAG........................ | 23 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAGATG..................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAGATGt.................... | 27 | t | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAGA....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAGATGA.................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TTTTTTTGAGGCCTtgct.......................................... | 18 | tgct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................AGGAAGTAGAGCAGCACAGC.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................CCTAGGAAGTAGAGCAGCACAGCAGATGt.................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TTTTTTTGAGGCCTAGGA.......................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................TAGGAAGTAGAGCAGCACAGCAGATGAC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GCAGATGACTCCACAGTAGATGCAGGA. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TCCGAAAAAAAAAATAATCAATCTTCGGATACGTGTTTGTGGGTAAAGCAGTTGCCACATGACCATTAGGACCTGGCTCCGACTCCCAGAACCCATGGAAAGGGTTAGACTGCAGGATGCTTTCTCCTCCAAACCTATTGCCTTGAACTACTCCTGAAGACATTTGGTAAAATTCTGTTAACTTTTAATTTTTTTTTGAGGCCTAGGAAGTAGAGCAGCACAGCAGATGACTCCACAGTAGATGCAGGAA ....................................................................................................((((((((..((.((((.......)))).))...)))..)))))......(((..(((((.(((.(((.((......)).)))...))).)))))..))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................CCAGAACCCATGGAAAact..................................................................................................................................................... | 19 | act | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |