
| Gene: Zbtb1 | ID: uc007nye.1_intron_0_0_chr12_77488127_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(6) PIWI.ip |
(3) PIWI.mut |
(19) TESTES |
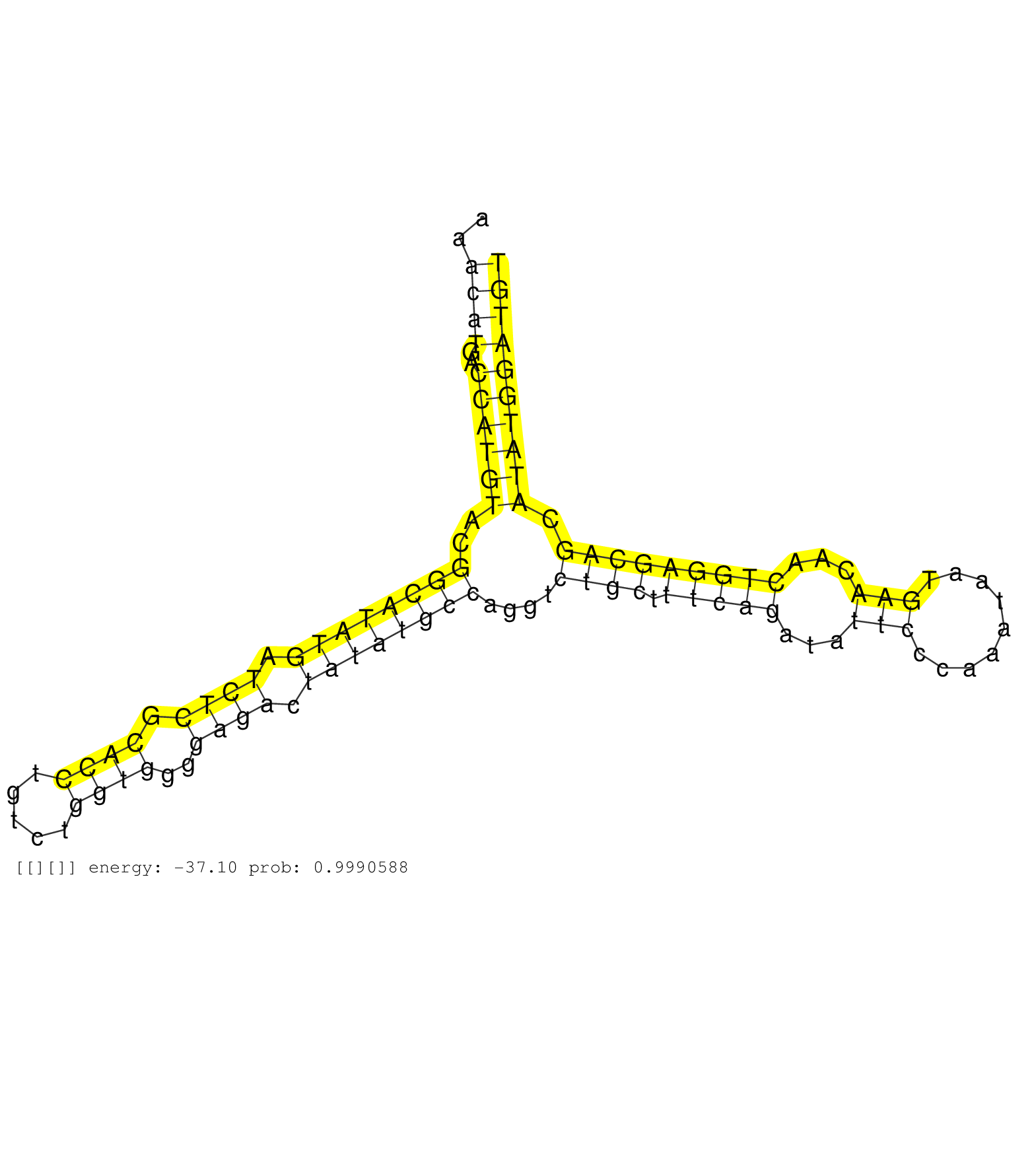
| AGAGAGCGCCAGTTGCGTCTACACAATGATATGCACAAAGGCATGGCCAGGTATGTCTGTTCCATTTGTGATCAAGGAAACTTCCGAAAACATGACCATGTACGGCATATGATCTCGCACCTGTCTGGTGGGGAGACTATATGCCAGGTCTGCTTTCAGATATTCCCAAATAATGAACAACTGGAGCAGCATATGGATGTCCATCTGTACACGTGTGGAATATGTGGAGCAAAGTTTAATTTGAGGAAGG .........................................................................................((((..((((((..((((((((.((((.((((.....))))..)))).))))))))....((((.(((((...(((.........)))...))))))))).)))))))))).................................................. .......................................................................................88..............................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................TGAACAACTGGAGCAGCATATGGATGTC................................................. | 28 | 1 | 12.00 | 12.00 | 5.00 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GTACACGTGTGGAATATGTGGAGC.................... | 24 | 1 | 7.00 | 7.00 | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGAACAACTGGAGCAGCATATGGATGT.................................................. | 27 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................AAATAATGAACAACTGGAGCAGCATA......................................................... | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................TAATGAACAACTGGAGCAGCATATGGA..................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TAATGAACAACTGGAGCAGCATATGGAT.................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGAACAACTGGAGCAGCATATGGATG................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................AAATAATGAACAACTGGAGCAGCATAT........................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGACCATGTACGGCATATGATCTCGCACC................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TATGGATGTCCATCTGTACACGTGTG................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GACCATGTACGGCATATGATCTCGCA................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CAAATAATGAACAACTGGAGCAGCATA......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGTGGAGCAAAGTTTAATTTGAGGAAGG | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TGGATGTCCATCTGTACACGTGTGGAA.............................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TCTGTACACGTGTGGAATATGTGGAG..................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TATGTGGAGCAAAGTTTAATTTGAGGAA.. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TATGGATGTCCATCTGTACACGTGTGG................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................ATAATGAACAACTGGAGCAa............................................................. | 20 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................ATAATGAACAACTGGAGCAGCA........................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATATGTGGAGCAAAGTTTAATTTGAGGA... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TATGATCTCGCACCTGTCTGGT......................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................AATAATGAACAACTGGAGCAGCA........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................CTGTCTGGTGGGGAGACTATATGCC......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................CCAAATAATGAACAACTGGAGCAGCA........................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGGAATATGTGGAGCAAAGTTTAATT......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AGAGAGCGCCAGTTGCGTCTACACAATGATATGCACAAAGGCATGGCCAGGTATGTCTGTTCCATTTGTGATCAAGGAAACTTCCGAAAACATGACCATGTACGGCATATGATCTCGCACCTGTCTGGTGGGGAGACTATATGCCAGGTCTGCTTTCAGATATTCCCAAATAATGAACAACTGGAGCAGCATATGGATGTCCATCTGTACACGTGTGGAATATGTGGAGCAAAGTTTAATTTGAGGAAGG .........................................................................................((((..((((((..((((((((.((((.((((.....))))..)))).))))))))....((((.(((((...(((.........)))...))))))))).)))))))))).................................................. .......................................................................................88..............................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................CAGGTCTGCTTTCAGATATTCCCAAATAAt............................................................................. | 30 | t | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GGTCTGCTTTCAGATATTCCCAAATAATGA.......................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....CGCCAGTTGCGTCTACACAATGATATGCAa....................................................................................................................................................................................................................... | 30 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................TGATCAAGGAAACTTCCGAAAACATGA........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCTGCTTTCAGATATTCCCAAATAATGA.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TCAGATATTCCCAtgta.................................................................................. | 17 | tgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....GCGCCAGTTGCGTCTACACAATGATATGCA....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |