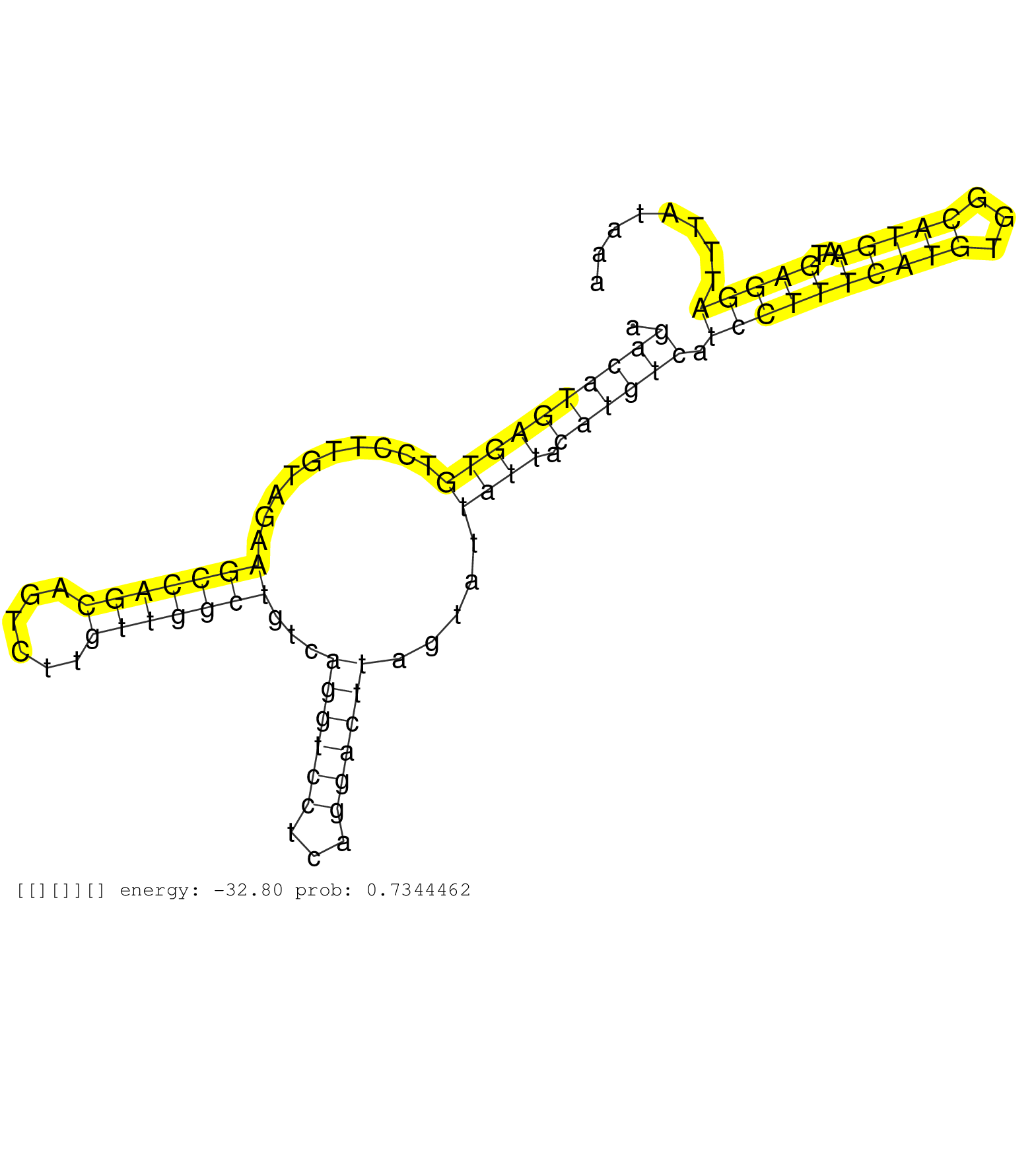
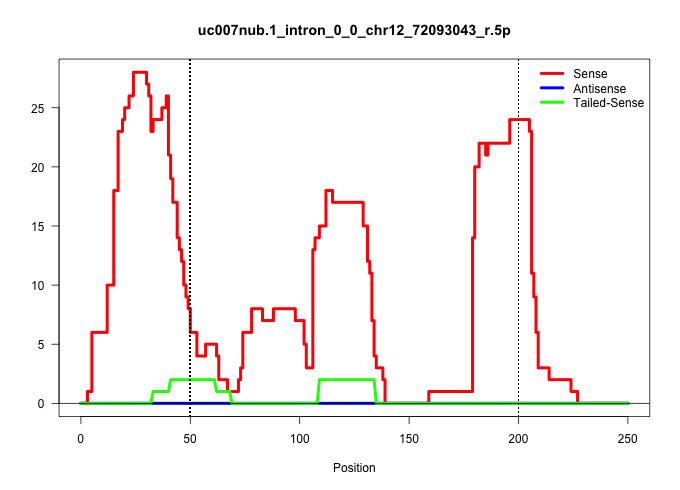
| Gene: AK015004 | ID: uc007nub.1_intron_0_0_chr12_72093043_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
| AATCTTGACTGGTGATGTGGAAAAAGAGACCCAGAAGAACAAAAAAACAGGTGACTGTAATGCTGGTCCCCCTGTGCAGGAACAGCTCAGGAGACTGTCGGAGACATGAGTGTCCTTGTAGAAGCCAGCAGTCTTGTTGGCTGTCAGGTCCTCAGGACTTAGTATTATTACATGTCATCCTTTCATGTGGCATGAATGAGGATTTATAAAGCCCTATTACGGTTTGGTTATGCAGTCCAAACAGACTTAC ......................................................................................................((((((((((..........(((((((......)))))))...((((((...)))))).....)))).)))))).((((((((((...)))))..)))))................................................ .....................................................................................................102.........................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................CTTTCATGTGGCATGAATGAGGATTTA............................................ | 27 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACTGGTGATGTGGAAAAAGAGACCC.......................................................................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | - | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............TGATGTGGAAAAAGAGACCCAGAAGAAC.................................................................................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGAGTGTCCTTGTAGAAGCCAGCAGTC..................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTTCATGTGGCATGAATGAGGATTTA............................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGAGTGTCCTTGTAGAAGCCAGCAG....................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGTGGAAAAAGAGACCCAGAAGAACAAAA.............................................................................................................................................................................................................. | 29 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTTTCATGTGGCATGAATGAGGATTTATAA......................................... | 30 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTTCATGTGGCATGAATGAGGATTTAT........................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TCCTTGTAGAAGCCAGCAGTCTTGTTG............................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTTTCATGTGGCATGAATGAGGATTTATA.......................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GTGTCCTTGTAGAAGCCAGCAGTCTa................................................................................................................... | 26 | a | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGTGGAAAAAGAGACCCAGAAGAACAA................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGAGTGTCCTTGTAGAAGCCAGCAGTCT.................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................AGAGACCCAGAAGAACAAAAAAACAGGTG..................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCAGGAACAGCTCAGGAGACTGTCGGA.................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGTGGAAAAAGAGACCCAGAAGAACA................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGGAAAAAGAGACCCAGAAGAACAAAAA............................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCAGGAACAGCTCAGGAGACTGTCGG..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................GGAACAGCTCAGGAGACTGTCGGAG................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGGAAAAAGAGACCCAGAAGAACAAAAAAA........................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGGAAAAAGAGACCCAGAAGAACAAAAAAAC.......................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTTCATGTGGCATGAATGAGGATTT............................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...CTTGACTGGTGATGTGGAAAAAGAGAC............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................GGAACAGCTCAGGAGACTGTCGGA.................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AGACATGAGTGTCCTTGTAGAAGCCAGC......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACTGGTGATGTGGAAAAAGAGACC........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGTGGAAAAAGAGACCCAGAAGAAC.................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGGAGACTGTCGGAGACATGAGTGTCC....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAAAAAGAGACCCAGAAGAACAAAAAAA........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGGAAAAAGAGACCCAGAAGAACAAAAAA............................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GAAGAACAAAAAAACAGGTGACTGTAAag............................................................................................................................................................................................ | 29 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................GTGGCATGAATGAGGATTTATAAAGCCC.................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................GAGTGTCCTTGTAGAAGCCAGCAGTCT.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GAAGAACAAAAAAACAGGTGACTGTAATG............................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................AAAAAACAGGTGACTGTAATGCTGGTCt..................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GACATGAGTGTCCTTGTAGAAGCCAGC......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AAAAAGAGACCCAGAAGAACAAAAAAACA......................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GGAGACATGAGTGTCaaa..................................................................................................................................... | 18 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................TCATGTGGCATGAATGAGGATTTATA.......................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTGCAGGAACAGCTCAGGAGACTGT........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................AGAAGAACAAAAAAACAG........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................AACAGGTGACTGTAATGCTGGT....................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGAGGATTTATAAAGCCCTATTACGGTTTGG....................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCATGTGGCATGAATGAGGATTTATAA......................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GTGTCCTTGTAGAAGCCAGCAGTCTT................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AAAGAGACCCAGAAGAACAAAAAAACAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGAGTGTCCTTGTAGAAGCCAGCAGT...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................AACAAAAAAACAGGTGACTGTAATGC........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TCCTTGTAGAAGCCAGCAGTCTTGTT................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GTGCAGGAACAGCTCAGGAGACTGTCGGAG................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................TAATGCTGGTCCCCCTGTGCAGGAAC....................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGAGGATTTATAAAGCCCTATTACGGTT.......................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TAGTATTATTACATGTCATCCTTTCA................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................CAAAAAAACAGGTGACTGTAATGC........................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| AATCTTGACTGGTGATGTGGAAAAAGAGACCCAGAAGAACAAAAAAACAGGTGACTGTAATGCTGGTCCCCCTGTGCAGGAACAGCTCAGGAGACTGTCGGAGACATGAGTGTCCTTGTAGAAGCCAGCAGTCTTGTTGGCTGTCAGGTCCTCAGGACTTAGTATTATTACATGTCATCCTTTCATGTGGCATGAATGAGGATTTATAAAGCCCTATTACGGTTTGGTTATGCAGTCCAAACAGACTTAC ......................................................................................................((((((((((..........(((((((......)))))))...((((((...)))))).....)))).)))))).((((((((((...)))))..)))))................................................ .....................................................................................................102.........................................................................................................210...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................CCCAGAAGAACAAAAagaa.............................................................................................................................................................................................................. | 19 | agaa | 4.00 | 0.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................GAAGAACAAAAAAAgggc........................................................................................................................................................................................................... | 18 | gggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ACCCAGAAGAACAAAgat............................................................................................................................................................................................................... | 18 | gat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CCCAGAAGAACAAAAagct.............................................................................................................................................................................................................. | 19 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................ACCCAGAAGAACAAAAga.............................................................................................................................................................................................................. | 18 | ga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................ACCCAGAAGAACAAgata................................................................................................................................................................................................................ | 18 | gata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CCCAGAAGAACAAAAAct............................................................................................................................................................................................................. | 18 | ct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |