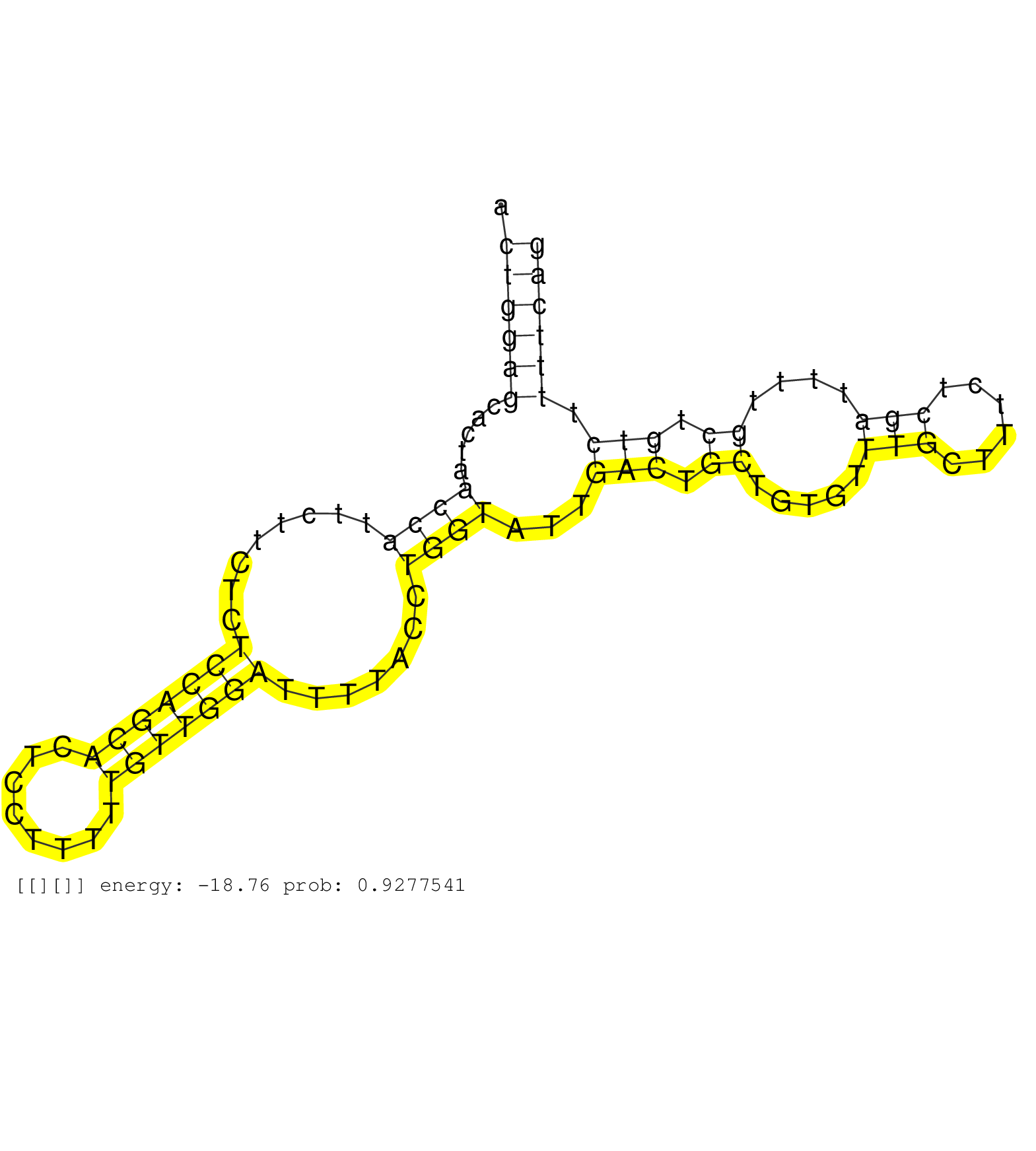
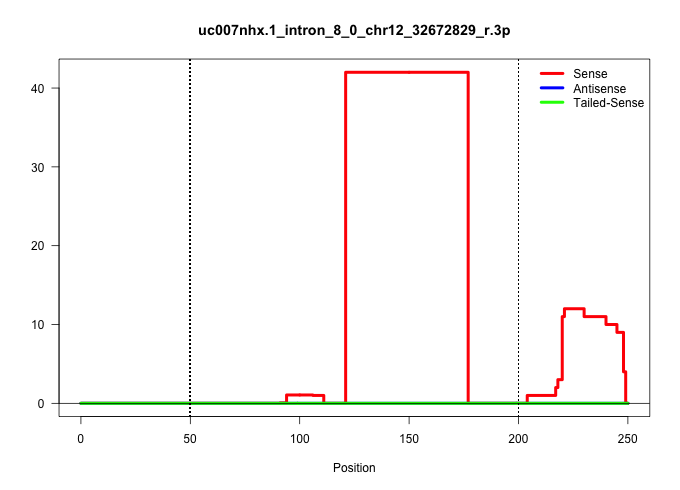
| Gene: Prkar2b | ID: uc007nhx.1_intron_8_0_chr12_32672829_r.3p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
| ATGGTCTTTCCACCCAGAAGTGGTGGTGTCATTGTCCTGTGCTCGTGGGCTAATCCTTGTTTGCCACCCAAAGCAAATGTCCTACCAACTGGGAGAGTGGACTGGAGCACTAACCATTCTTCTCTCCAGCACTCCTTTTTGTTGGATTTTACCTGGTATTGACTGCTGTGTTTGCTTTCTCGATTTTGCTGTCTTTTCAGATAATACATCCCAAAACCGATGATCAGAGAAACAGATTGCAAGAGGCTTG .....................................................................................................((((((.....((((........(((((((........))))))).......))))...(((.((.....(((......)))....)).))).)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CTCTCCAGCACTCCTTTTTGTTGGATTTTACCTGGTATTGACTGCTGTGTTT............................................................................. | 52 | 1 | 42.00 | 42.00 | 42.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATCAGAGAAACAGATTGCAAGAGGCT.. | 28 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................................................................................................TGATCAGAGAAACAGATTGCAAGAGGCTT. | 29 | 1 | 4.00 | 4.00 | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................GATGATCAGAGAAACAGATTGCAAGAGGCT.. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................GAGTGGACTGGAGCACT........................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TACATCCCAAAACCGATGATCAGAGA.................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................CGATGATCAGAGAAACAGATTGC.......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................................GATCAGAGAAACAGATTGCAAGAG..... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................GGAGAGTGGACTGGA................................................................................................................................................ | 15 | 17 | 0.06 | 0.06 | - | - | - | 0.06 | - | - | - | - | - | - |
| ATGGTCTTTCCACCCAGAAGTGGTGGTGTCATTGTCCTGTGCTCGTGGGCTAATCCTTGTTTGCCACCCAAAGCAAATGTCCTACCAACTGGGAGAGTGGACTGGAGCACTAACCATTCTTCTCTCCAGCACTCCTTTTTGTTGGATTTTACCTGGTATTGACTGCTGTGTTTGCTTTCTCGATTTTGCTGTCTTTTCAGATAATACATCCCAAAACCGATGATCAGAGAAACAGATTGCAAGAGGCTTG .....................................................................................................((((((.....((((........(((((((........))))))).......))))...(((.((.....(((......)))....)).))).)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................TTCAGATAATACATCc........................................ | 16 | c | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |