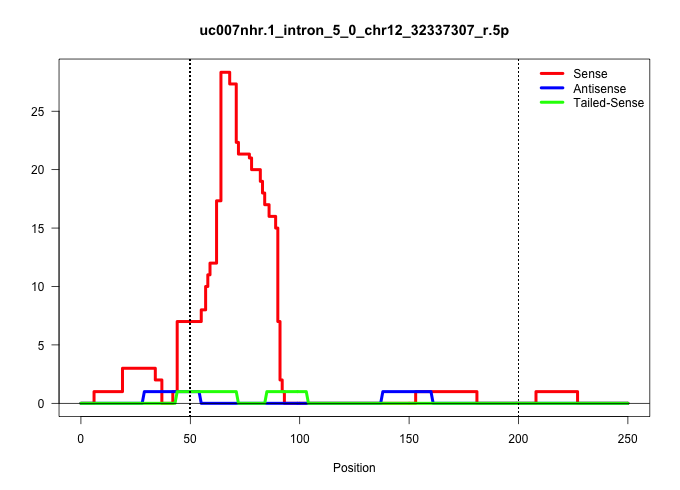
| Gene: Dus4l | ID: uc007nhr.1_intron_5_0_chr12_32337307_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(12) TESTES |
| GGGCCTTCTCCCATTCCGCTCCGGACGCAGCCACCGCGCCGATCTCCAACGCAAGTATTGGGCCTGAGAACTCCGGCGTGGGGAAGAGGCTCTTGGGAGCGTCACGGCCCTCCTCGCCCAACACGTGGGAGCTTAGGGCTGCTCAGCCTTACCTAGTCCCCGCCTGGAGTGGCAAGCCCGGGTAGCAGGCAGTATATGCTGGGGAAGAATAGTTGTTAAGCTCTGCCCTTAAAAATAAAAACGTTTAAAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TGAGAACTCCGGCGTGGGGAAGAGGCT............................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ............................................TCCAACGCAAGTATTGGGCCTGAGAAC................................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTGAGAACTCCGGCGTGGGGAAGAGGC................................................................................................................................................................ | 28 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGAGAACTCCGGCGTGGGGAAGAGGC................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................TCCGGACGCAGCCACCGC..................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| .........................................................TTGGGCCTGAGAACTCCGGCGTGGG........................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......TCTCCCATTCCGCTCCGGACGCAGCCAC........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................TGAGAACTCCGGCGTGGGGAAGAGGCTCT............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGAGAACTCCGGCGTGGGGAAGAGG................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TATTGGGCCTGAGAACTCCGGCGTGGGG....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................TCTCCAACGCAAGTATTGGGCCTGAG...................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ATAGTTGTTAAGCTCTGCC....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................TAGTCCCCGCCTGGAGTGGCAAGCCCGG..................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................TCCAACGCAAGTATTGGGCCTGAGAAat.................................................................................................................................................................................. | 28 | at | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TCCAACGCAAGTATTGGGCCTGAGAACT.................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................GAGGCTCTTGGGAGCtttt.................................................................................................................................................. | 19 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................TGGGCCTGAGAACTCCGGCGTGGGGAAG.................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TTGGGCCTGAGAACTCCGGCGTGGGGA...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGCCTGAGAACTCCGGCG............................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................TGAGAACTCCGGCGTGGGGAAGAGGCTC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTGAGAACTCCGGC............................................................................................................................................................................. | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GGGCCTTCTCCCATTCCGCTCCGGACGCAGCCACCGCGCCGATCTCCAACGCAAGTATTGGGCCTGAGAACTCCGGCGTGGGGAAGAGGCTCTTGGGAGCGTCACGGCCCTCCTCGCCCAACACGTGGGAGCTTAGGGCTGCTCAGCCTTACCTAGTCCCCGCCTGGAGTGGCAAGCCCGGGTAGCAGGCAGTATATGCTGGGGAAGAATAGTTGTTAAGCTCTGCCCTTAAAAATAAAAACGTTTAAAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................CTGCTCAGCCTTACCTAGTCCCC......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................GCCACCGCGCCGATCTCCAACGCAAG................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |