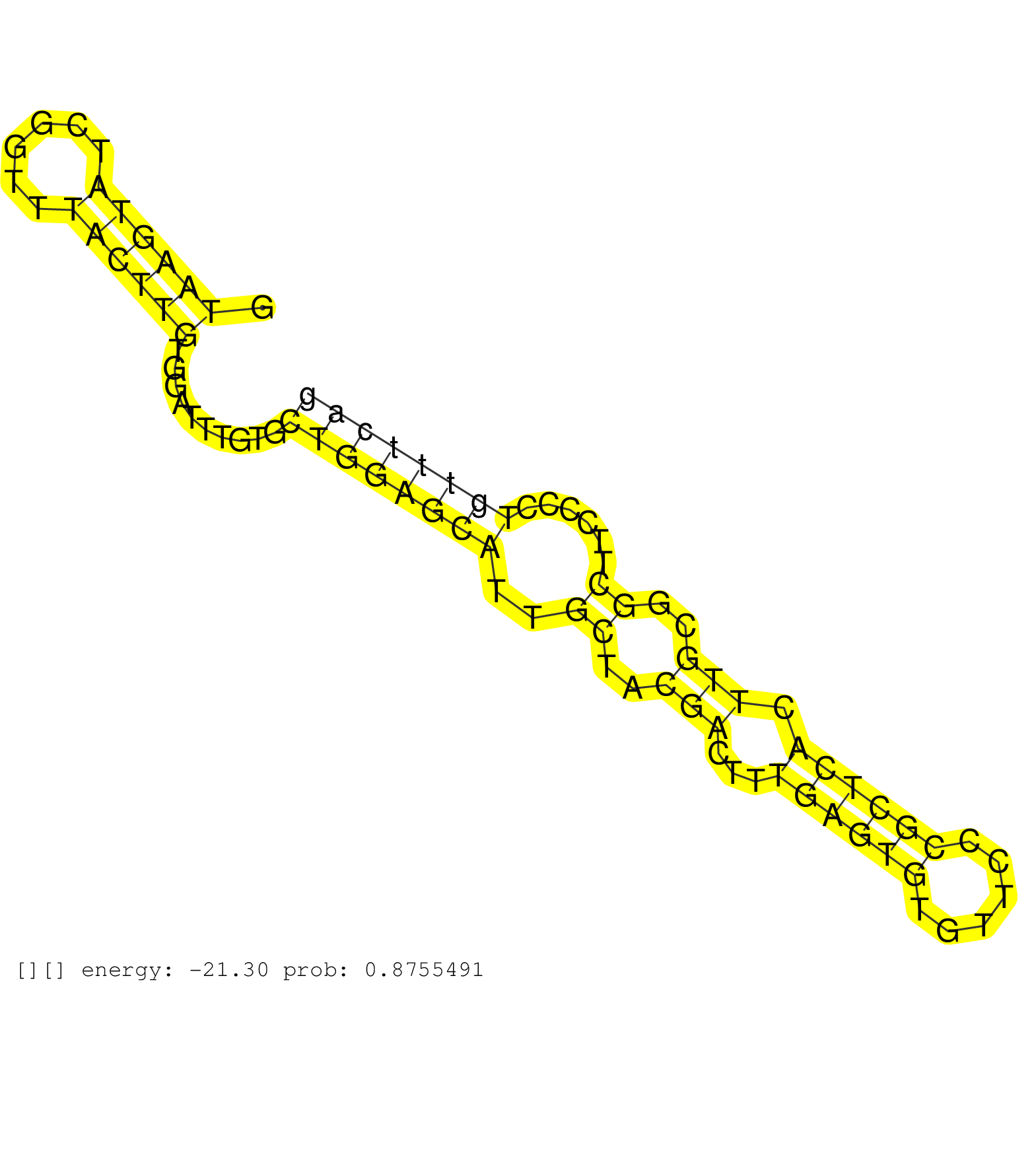
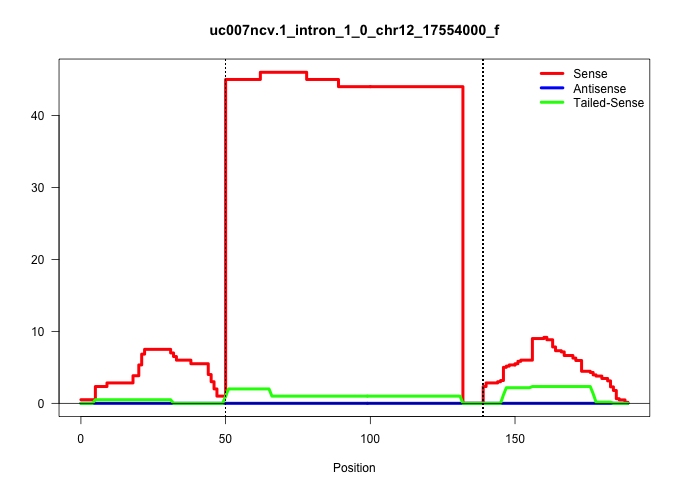
| Gene: Odc1 | ID: uc007ncv.1_intron_1_0_chr12_17554000_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(7) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(3) PIWI.mut |
(35) TESTES |
| GACCTTGTGAGGAGCTGGTGATAATTTGATTCCATCTCCAGGTTCCCTGTGTAAGTATCGGTTTACTTGTGGATTTGTGCTGGAGCATTGCTACGACTTTGAGTGTGTTCCCGCTCACTTGCGGCTTCCCCTGTTTCAGAAGCACATCGAGAACCATGAGCAGCTTTACTAAGGACGAGTTTGACTGCC ...................................................((((((......))))))..........((((((((..((..(((...((((((......)))))).)))..))......)))))))).................................................. ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTATCGGTTTACTTGTGGATTTGTGCTGGAGCATTGCTACGACTTTGA....................................................................................... | 52 | 1 | 55.00 | 55.00 | 55.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTTTACTAAG................ | 27 | 6 | 1.33 | 1.33 | - | 0.50 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| ..................TGATAATTTGATTCCATCTCCAGGTT................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAGCACATCGAGAACCATGAGCAG.......................... | 24 | 2 | 1.00 | 1.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTTTACTAAGGAat............ | 31 | at | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTTG...... | 27 | 6 | 1.00 | 1.00 | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | - | - | - | - |
| ....................ATAATTTGATTCCATCTCCAGGTTC................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTATCGGTTTACTTGTGGATTTGT............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AATTTGATTCCATCTCCAGGTTCC............................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTATCGGTcctg........................................................................................................................... | 15 | cctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CGAGAACCATGAGCAGCTTTACTAAGGACGt........... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TAATTTGATTCCATCTCCAGGTTCCCTGT........................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TAATTTGATTCCATCTCCAGGTTCCC.............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ACATCGAGAACCATGAG............................. | 17 | 7 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TTACTTGTGGATTTGTGCTGGAGCATT.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTTGAC.... | 29 | 6 | 1.00 | 1.00 | - | 0.17 | - | 0.17 | - | 0.17 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| .....TGTGAGGAGCTGGTGATAATTTGATTC............................................................................................................................................................. | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAGGAGCTGGTGATAATTTGATTa............................................................................................................................................................. | 27 | a | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........AGGAGCTGGTGATAATTTGATTCCATCTC....................................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAGGAGCTGGTGATAATTTGATTCC............................................................................................................................................................ | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAGCACATCGAGAACCATGAGCAGC......................... | 25 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAGCACATCGAGAACCATGAGCAGCTTT...................... | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAGGAGCTGGTGATAATTTGATT.............................................................................................................................................................. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................ATAATTTGATTCCATCTCCAGGTT................................................................................................................................................. | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGCACATCGAGAACCATGAGCAGCTTTACT................... | 30 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTTGA..... | 28 | 6 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - |
| .......................................................................................................................................................................ACTAAGGACGAGTTTGACT... | 19 | 7 | 0.43 | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.43 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAGCACATCGAGAACCATGAGC............................ | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGTGAGGAGCTGGTGAT....................................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTTGACTGC. | 32 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTT....... | 26 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TAAGGACGAGTTTGAC.... | 16 | 7 | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CGAGAACCATGAGCAGCTTTACTAAG................ | 26 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| ................................................................................................................................................CATCGAGAACCATGAGCAGCTTTACTA.................. | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - |
| ......................................................................................................................................................GAACCATGAGCAGCTTTACTAAGGACGAGT......... | 30 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGAACCATGAGCAGCTTTACTAAGGACG............ | 29 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AACCATGAGCAGCTTTACTAAGGACGA........... | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CAGCTTTACTAAGGACGAGTTTGACT... | 26 | 6 | 0.17 | 0.17 | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................ACCATGAGCAGCTTTACTAAGGACG............ | 25 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CGAGAACCATGAGCAGCTTTACTAAGG............... | 27 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTTTACTAAGGAC............. | 30 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTTTACTAAGGACtt........... | 32 | tt | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CCATGAGCAGCTTTACTAAGGACGAGTT........ | 28 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |
| .......................................................................................................................................................AACCATGAGCAGCTTTACTAAGGACGAGT......... | 29 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTT....................... | 20 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCGAGAACCATGAGCAGCTTTACTA.................. | 25 | 6 | 0.17 | 0.17 | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ATCGAGAACCATGAGCAGC......................... | 19 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TGAGCAGCTTTACTAAGGACGAGTTTGt..... | 28 | t | 0.17 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................AAGGACGAGTTTGACTGCC | 19 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTTACTAAGGACGAGTTTGAC.... | 21 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GACCTTGTGAGGAGCTGGTGATAATTTGATTCCATCTCCAGGTTCCCTGTGTAAGTATCGGTTTACTTGTGGATTTGTGCTGGAGCATTGCTACGACTTTGAGTGTGTTCCCGCTCACTTGCGGCTTCCCCTGTTTCAGAAGCACATCGAGAACCATGAGCAGCTTTACTAAGGACGAGTTTGACTGCC ...................................................((((((......))))))..........((((((((..((..(((...((((((......)))))).)))..))......)))))))).................................................. ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|