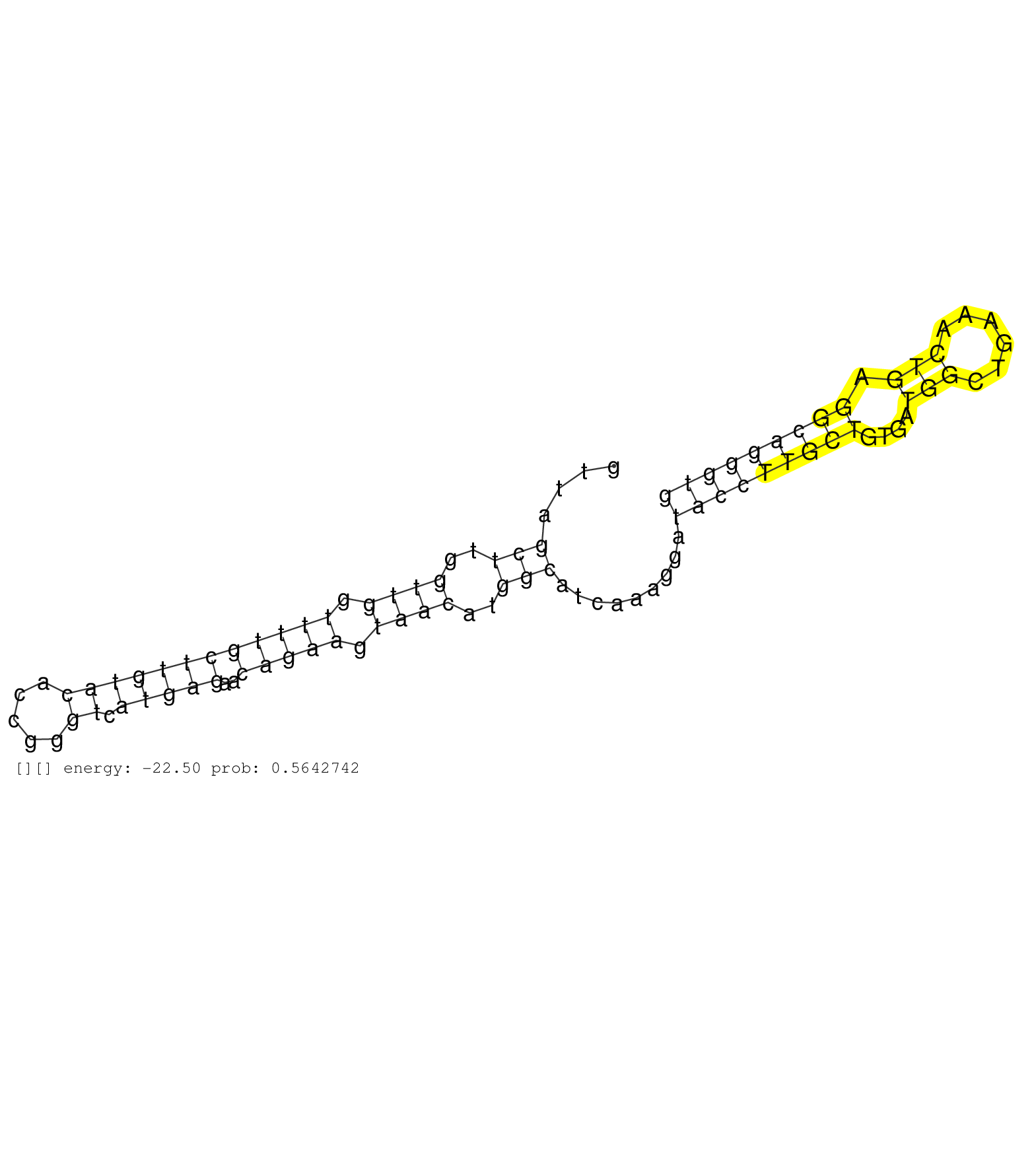

| Gene: E2f6 | ID: uc007ncc.1_intron_3_0_chr12_16823261_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| AAAAATAAGGATTAATCTAGAAGAAAATGTACAGTATGTGTCCATGAGAAGTAAGTTGATTCCTAAGTGAGTTTGGGTTTTCTGGTTTGGGCATCACGTGGTTAGCTTGGTTGGTTTTGCTTGTACACCGGGTCATGAGAACAGAAGTAACATGGCATCAAAGGATACCTTGCTGTGATGGCTGAAACTGAGGCAGGGTGGGAGGAACTGATGTCTTTGCCCCCTTAGCGCAGTGGCAGCACTAAACATG ........................................................................................................(((..((((.((((((((((((.....)).)))))..))))).))))..))).........(((((((((....(((......))).))))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGG......................................................... | 24 | 1 | 45.00 | 45.00 | 14.00 | 9.00 | - | 2.00 | 4.00 | 1.00 | 7.00 | 1.00 | 5.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGG......................................................... | 23 | 1 | 41.00 | 41.00 | 13.00 | 8.00 | - | 6.00 | - | 6.00 | - | 4.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGa......................................................... | 23 | a | 11.00 | 2.00 | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGGCAGG..................................................... | 28 | 1 | 6.00 | 6.00 | - | - | - | - | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGGC........................................................ | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....TAAGGATTAATCTAGAAGAAAATGTACAG........................................................................................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGGCAG...................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGa......................................................... | 24 | a | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............ATCTAGAAGAAAATGTACAGTATGTG.................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGGCAGGGTGG................................................. | 32 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAG.......................................................... | 22 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CTTGCTGTGATGGCTGAAACTGAGG......................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAag......................................................... | 24 | ag | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGGCAGGG.................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGGCAGGa.................................................... | 29 | a | 1.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...AATAAGGATTAATCTAGAAGAAAATG............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....TAAGGATTAATCTAGAAGAAAATG............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GCTGTGATGGCTGAAACTGAGG......................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ATAAGGATTAATCTAGAAGAAAATGTACAGT....................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGGCAGG..................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AAATAAGGATTAATCTAGAAGAAAATGTA........................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGAGGCAG...................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TAAGGATTAATCTAGAAGAAAATGTA........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .AAAATAAGGATTAATCTAGAAG................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGCTGTGATGGCTGAAACTGAGGCAGaa.................................................... | 28 | aa | 1.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TAGAAGAAAATGTACAGTATGTGTCCA.............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TTGCTGTGATGGCTGAAACTGA........................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAAAATAAGGATTAATCTAGAAGAAAATGTACAGTATGTGTCCATGAGAAGTAAGTTGATTCCTAAGTGAGTTTGGGTTTTCTGGTTTGGGCATCACGTGGTTAGCTTGGTTGGTTTTGCTTGTACACCGGGTCATGAGAACAGAAGTAACATGGCATCAAAGGATACCTTGCTGTGATGGCTGAAACTGAGGCAGGGTGGGAGGAACTGATGTCTTTGCCCCCTTAGCGCAGTGGCAGCACTAAACATG ........................................................................................................(((..((((.((((((((((((.....)).)))))..))))).))))..))).........(((((((((....(((......))).))))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|