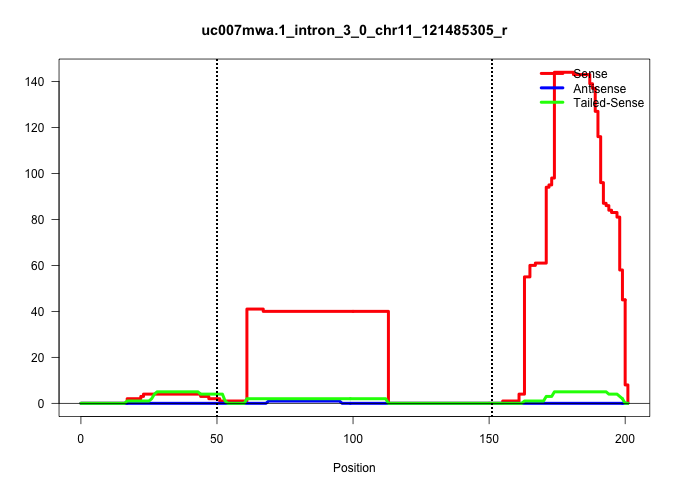
| Gene: B3gntl1 | ID: uc007mwa.1_intron_3_0_chr11_121485305_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
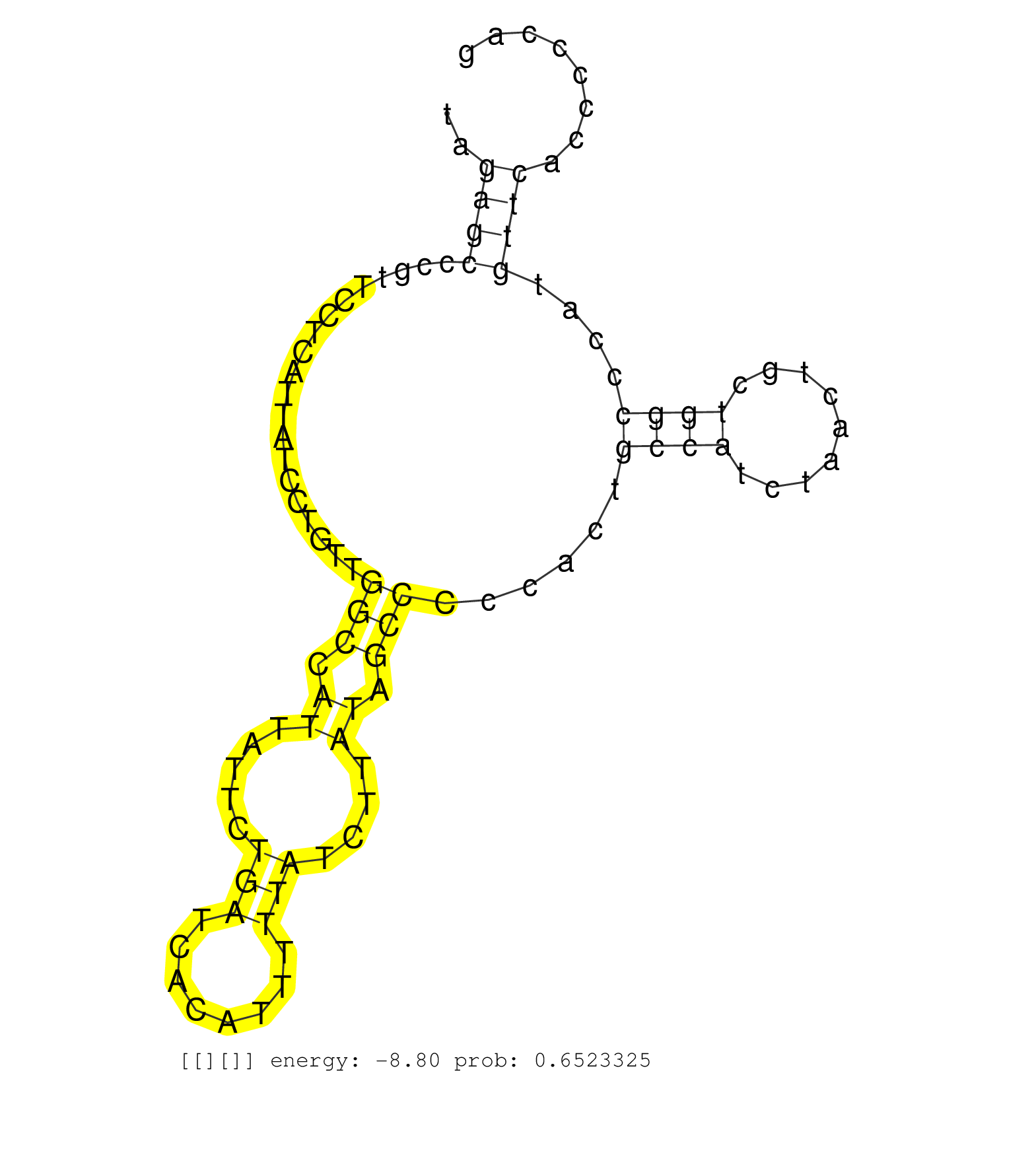
| AGCAGGGGCGTAAGTTATACCGCAGCCTGACTGCAGCCAGCCGGCACAAGGTAGAGCCCGTTCCTCATTATCCTGTTGGCCATTATTCTGATCACATTTTTATCTTATAGCCCCCACTGCCATCTAACTGCTGGCCCATGTTCACCCCCAGGTGGTGGCCTTCTGTGACGTTGATGAGAACAAGATCAGGAAGGGCTTCTA .....................................................((((....................(((.((.....(((........)))....)).)))......((((.........))))....)))).......................................................... ...................................................52.................................................................................................151................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................TCCTCATTATCCTGTTGGCCATTATTCTGATCACATTTTTATCTTATAGCCC........................................................................................ | 52 | 1 | 40.00 | 40.00 | - | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACAAGATCAGGAAGGGCTTCT. | 26 | 1 | 36.00 | 36.00 | 12.00 | - | 9.00 | 8.00 | 1.00 | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGAGAACAAGATCAGGAAGGGCTT... | 27 | 1 | 21.00 | 21.00 | 5.00 | - | 3.00 | 2.00 | 5.00 | - | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGA.......... | 28 | 1 | 19.00 | 19.00 | 8.00 | - | 1.00 | 4.00 | - | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGG........... | 27 | 1 | 11.00 | 11.00 | 1.00 | - | - | - | - | 6.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGAGAACAAGATCAGGAAGGGCTTC.. | 28 | 1 | 10.00 | 10.00 | 6.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAG............ | 26 | 1 | 10.00 | 10.00 | - | - | - | - | 2.00 | 4.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACAAGATCAGGAAGGGCTTCTA | 27 | 1 | 8.00 | 8.00 | 1.00 | - | 3.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGAA......... | 29 | 1 | 6.00 | 6.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGACGTTGATGAGAACAAGATCAGGAA......... | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TCTGTGACGTTGATGAGAACAAGATC.............. | 26 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................ATGAGAACAAGATCAGGAAGGGCTTC.. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTGATGAGAACAAGATCAGGAA......... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACAAGATCAGGAAGGGCTTta. | 26 | ta | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGAGAACAAGATCAGGAAGGGCT.... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCA............. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GACTGCAGCCAGCCGGCACAAGGTg.................................................................................................................................................... | 25 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................ATGAGAACAAGATCAGGAAGGGCTTCT. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TACCGCAGCCTGACTGCAGCCAGCCGGCAC.......................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGAAGt....... | 31 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CTGACTGCAGCCAGCCGGCACAAGGTg.................................................................................................................................................... | 27 | g | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GATGAGAACAAGATCAGGAAGGGCTT... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATC.............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGACGTTGATGAGAACAAGATCAGGAAG........ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGAGAACAAGATCAGGAAGGGCTa... | 27 | a | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGCCTGACTGCAGCCAGCCGGCACAAGG...................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGATGAGAACAAGATCAGGAAGGGCgtc.. | 28 | gtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGACGTTGATGAGAACAAGATCAGGA.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TACCGCAGCCTGACTGCAGCCAGCCGG............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGAGCCCGTTCCTCA...................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGACTGCAGCCAGCCGGCACAAGGTAt................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CAGCCTGACTGCAGCCAGCCGGCACAAG....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGACTGCAGCCAGCCGGCACAAGGTg.................................................................................................................................................... | 26 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGAAGGG...... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................ACGTTGATGAGAACAAGATCAGGAAGG....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGGCCTTCTGTGACGTTGATGAGAAC.................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACAAGATCAGGAAGGGCTTC.. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAACAAGATCAGGAAGGGCTT... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TACCGCAGCCTGACTGCAGCCAGCCGt............................................................................................................................................................. | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGAAGGGC..... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................TGTGACGTTGATGAGAACAAGATCAGGAAGG....... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCAGGGGCGTAAGTTATACCGCAGCCTGACTGCAGCCAGCCGGCACAAGGTAGAGCCCGTTCCTCATTATCCTGTTGGCCATTATTCTGATCACATTTTTATCTTATAGCCCCCACTGCCATCTAACTGCTGGCCCATGTTCACCCCCAGGTGGTGGCCTTCTGTGACGTTGATGAGAACAAGATCAGGAAGGGCTTCTA .....................................................((((....................(((.((.....(((........)))....)).)))......((((.........))))....)))).......................................................... ...................................................52.................................................................................................151................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................TGCAGCCAGCCGGCAtctc........................................................................................................................................................... | 19 | tctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................ATCCTGTTGGCCATTATTCTGATCACA......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGCAGCCAGCCGGCAtccc........................................................................................................................................................... | 19 | tccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................AGCCTGACTGCAGCCAat.................................................................................................................................................................. | 18 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................TGCAGCCAGCCGGCACAAtc........................................................................................................................................................ | 20 | tc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |