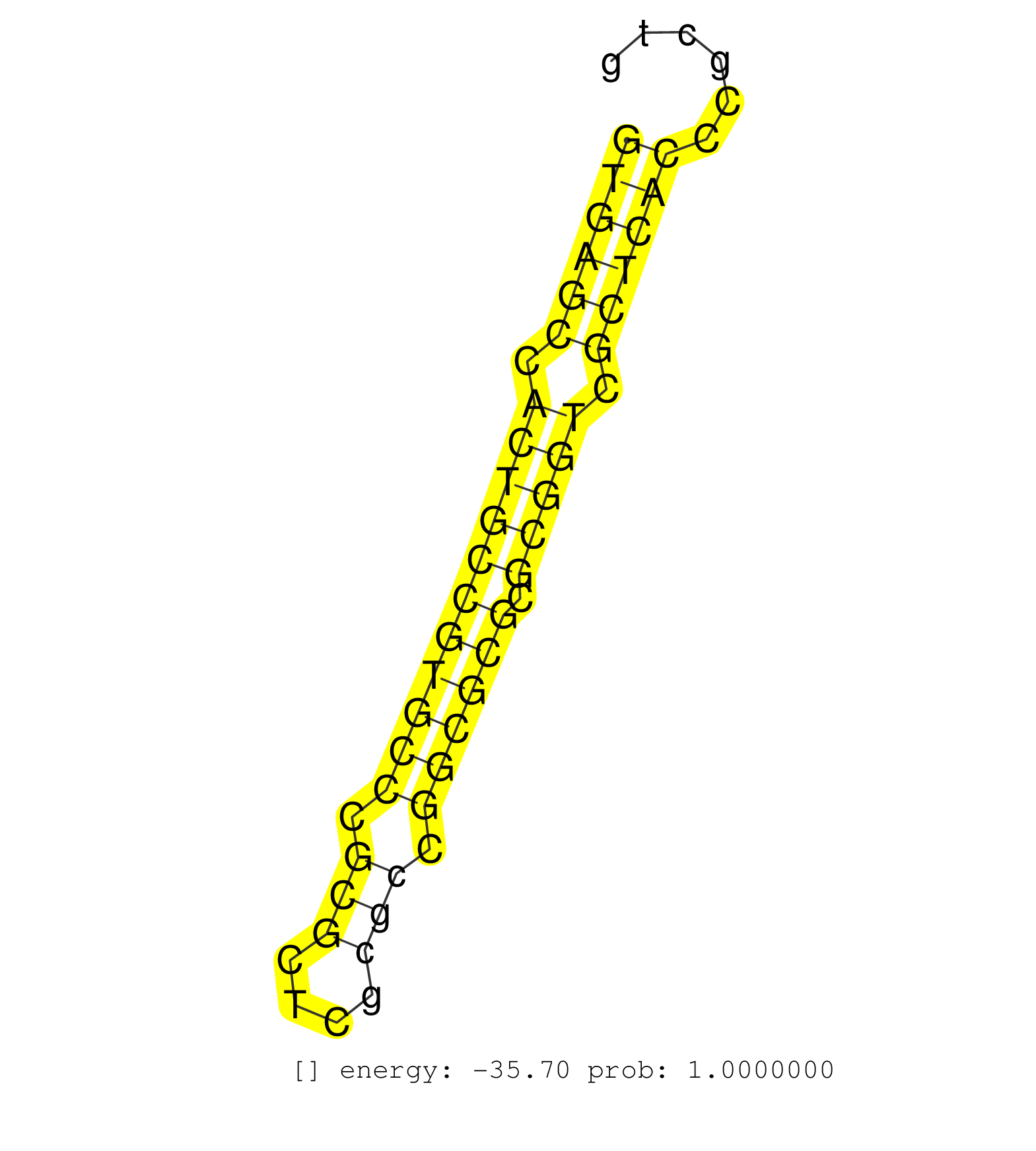

| Gene: Sirt7 | ID: uc007mtr.1_intron_8_0_chr11_120486125_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| AGCGGGTCCGGAGGCTGCGGGAGGAGCAGCAGCGGGAGCGCCTCCGCCAGGTGAGCCACTGCCGTGCCCGCGCTCGCGCCGGCGCGCGCGGTCGCTCACCCGCTGCTCGTCCGTAGGTGTCACGCATCCTGAGGAAGGCGGCTGCAGAGCGCAGCGCGGAGGAGGG ..................................................((((((.(((((((((((.(((....))).)))))).))))).))))))................................................................... ..................................................51....................................................105........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................CGGCGCGCGCGGTCGCTCACCC................................................................. | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................CGGCGCGCGCGGTCGCTCACC.................................................................. | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TAGGTGTCACGCATCCTGAGGAAGGC........................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCTGAGGAAGGCGGCTGCAGAGCGCAG............ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TCACCCGCTGCTCGTCCGTAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................CGGCGCGCGCGGTCGCTCACCCa................................................................ | 23 | a | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CGGCGCGCGCGGTCGCTCAC................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................CAGCAGCGGGAGCGCCTCCGCCAG.................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGCGGGAGGAGCAGgtgg..................................................................................................................................... | 18 | gtgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................CGGCGCGCGCGGTCGCTCACCCG................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCACTGCCGTGCCCGCttat........................................................................................... | 25 | ttat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCTGAGGAAGGCGGCTGCAGAGCGCA............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGCCACTGCCGTGCCCG................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................AGGCGGCTGCAGAGCGCAGCGCGGt...... | 25 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCTGAGGAAGGCGGCTGCAGAGCGC.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................TCCTGAGGAAGGCGGCTGCAGAGCGt.............. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................CGGCGCGCGCGGTCG........................................................................ | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....GTCCGGAGGCTGCGG.................................................................................................................................................. | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| AGCGGGTCCGGAGGCTGCGGGAGGAGCAGCAGCGGGAGCGCCTCCGCCAGGTGAGCCACTGCCGTGCCCGCGCTCGCGCCGGCGCGCGCGGTCGCTCACCCGCTGCTCGTCCGTAGGTGTCACGCATCCTGAGGAAGGCGGCTGCAGAGCGCAGCGCGGAGGAGGG ..................................................((((((.(((((((((((.(((....))).)))))).))))).))))))................................................................... ..................................................51....................................................105........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................GCTCGTCCGTAGGTGTCACGCATCCTGA.................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................GCTGCTCGTCCGTAGGTGTCACGCA........................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GTCGCTCACCCGCTGCTCGTCCGTA................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |