
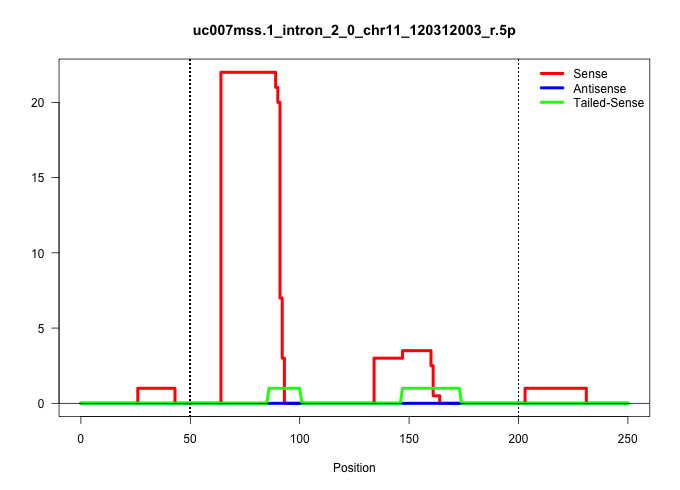
| Gene: Pde6g | ID: uc007mss.1_intron_2_0_chr11_120312003_r.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(11) TESTES |
| AGGTCCAGATAACCGGCGCCTGTGGCTTCCTGAGATCTGTCCAGTGCTTGGTGAGTATGGCCCATGCTGTGGTCCTTTCTGACAGGTAGGCATTAGCCTGGTCCTGGTTCCTAGGCTGGTGCTGGCTTCAGGCATAGAGGCTGAATCTAGGAATGACAGAGGGGAGCCACTAGACTGGGGGGGAGCTCCCCCAGTTGTTGTTATTGCCTACAACAGACTTAATGCATCCCTGTTGCTTAGTCTCTCAGCT ............................................................(((...((((.((((..(((((..(((.(((((((((((((.........))))))))))))).))))))))..((((.......)))))).)).))))..)))...................................................................................... ...........................................................60.......................................................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TGCTGTGGTCCTTTCTGACAGGTAGGC............................................................................................................................................................... | 27 | 1 | 13.00 | 13.00 | 8.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ................................................................TGCTGTGGTCCTTTCTGACAGGTAGGCA.............................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - |
| ................................................................TGCTGTGGTCCTTTCTGACAGGTAGGCAT............................................................................................................................................................. | 29 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................TAGAGGCTGAATCTAGGAATGACAGAG......................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TTGCCTACAACAGACTTAATGCATCCCT................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................TAGGCATTAGCCggt..................................................................................................................................................... | 15 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................TAGGAATGACAGAGGGGAGCCACTAGt............................................................................ | 27 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................TGCTGTGGTCCTTTCTGACAGGTAG................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................TTCCTGAGATCTGTCCA............................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................TAGAGGCTGAATCTAGGAATGACAGA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................TGCTGTGGTCCTTTCTGACAGGTAGG................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGGAATGACAGAGGGG...................................................................................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - |
| AGGTCCAGATAACCGGCGCCTGTGGCTTCCTGAGATCTGTCCAGTGCTTGGTGAGTATGGCCCATGCTGTGGTCCTTTCTGACAGGTAGGCATTAGCCTGGTCCTGGTTCCTAGGCTGGTGCTGGCTTCAGGCATAGAGGCTGAATCTAGGAATGACAGAGGGGAGCCACTAGACTGGGGGGGAGCTCCCCCAGTTGTTGTTATTGCCTACAACAGACTTAATGCATCCCTGTTGCTTAGTCTCTCAGCT ............................................................(((...((((.((((..(((((..(((.(((((((((((((.........))))))))))))).))))))))..((((.......)))))).)).))))..)))...................................................................................... ...........................................................60.......................................................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................GCCCATGCTGTGGTCCTTTCTGACAGGTA.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |