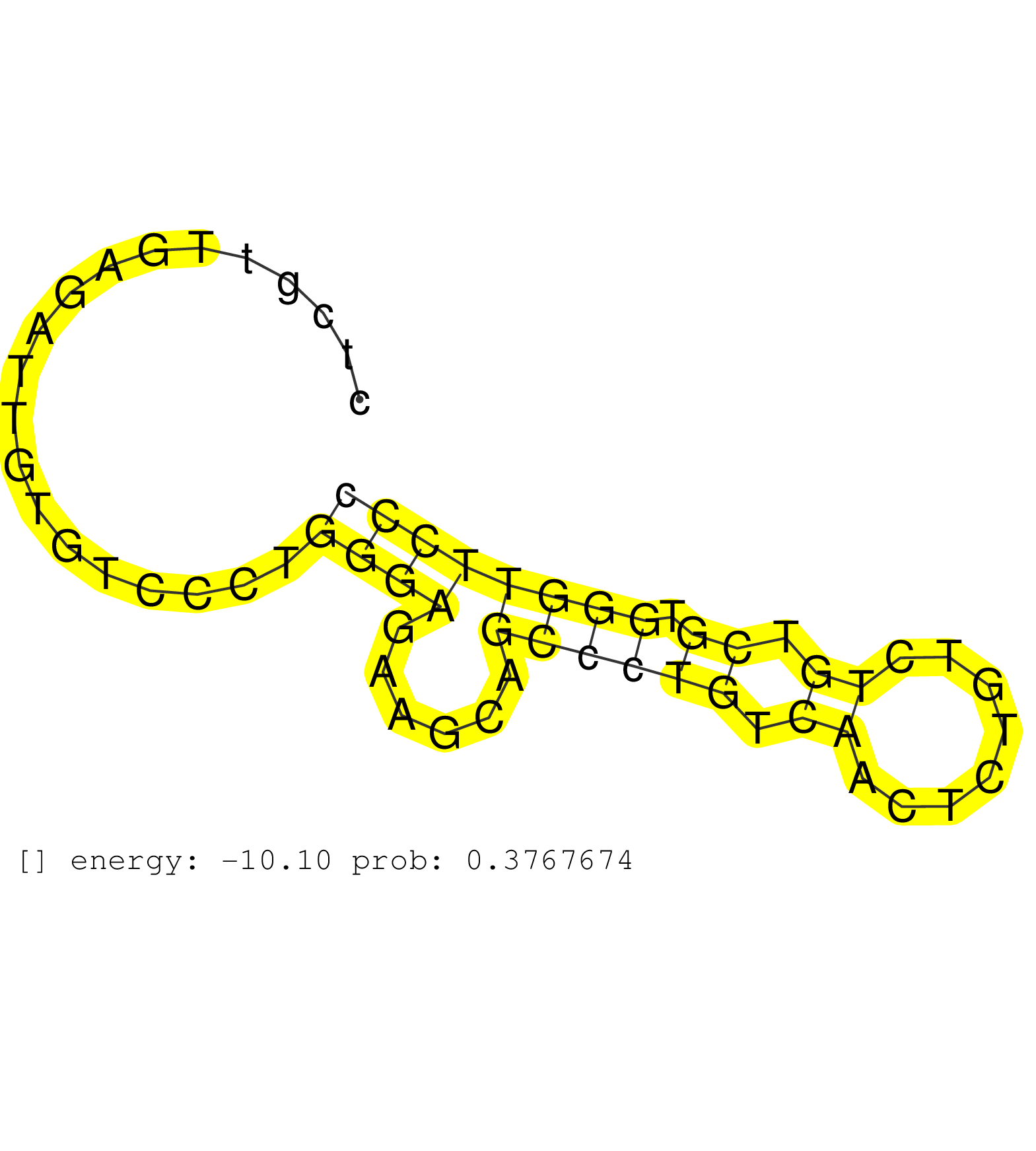

| Gene: 2310003H01Rik | ID: uc007msm.1_intron_3_0_chr11_120233541_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(18) TESTES |
| TCCAGGGAATAGCCCCAGATGGAACTGATGTCAACCTCACCGTCCATGAGGCAAGCAGGGGCCACTTGTCTGTTCTTGGGGAGACGGACGTCAGGGCTCATGGTCCTCAGTGAAGAGTTACTTTAACTGGCCTGTCAGTCTCGTTGAGATTGTGTCCCTGGGAGAAGCAGCCCTGTCAACTCTGTCTGTCGTGGGTTCCCGTTCTTGTGCACTGCTAGGGTTGCAGTGGACAGTGAATGCAGAAACCTCC ...............................................................................................................................................................((((......((((((.((........)).)).)))))))).................................................. ...........................................................................................................................................140.........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TGAGATTGTGTCCCTGGGAGAAGCAGC............................................................................... | 27 | 1 | 9.00 | 9.00 | 5.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................................GTCGTGGGTTCCCGTgtcg............................................ | 19 | gtcg | 9.00 | 0.00 | - | - | - | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGAGATTGTGTCCCTGGGAGAAGCAGCC.............................................................................. | 28 | 1 | 7.00 | 7.00 | 1.00 | - | 2.00 | - | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................TGAGATTGTGTCCCTGGGAGAAGCAG................................................................................ | 26 | 1 | 7.00 | 7.00 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................TGCAGTGGACAGTGAATGCAGAAACCTC. | 28 | 1 | 6.00 | 6.00 | - | - | 3.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TAGGGTTGCAGTGGACAGTGAATGC.......... | 25 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGTCAACTCTGTCTGTCGTGGGTTCC................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................TGAGATTGTGTCCCTGGGAGAAGCAGCCC............................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CTGTTCTTGGGGAGACGGACGTCAGG........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................CAGTGAATGCAGAAACCT.. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TAGGGTTGCAGTGGACAGTGAATGCAG........ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GATTGTGTCCCTGGGAGAAGCAGCCCT............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TCTTGGGGAGACGGACGTCAGGGCTC....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCAGTGGACAGTGAATGCAGAAACCT.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTGCAGTGGACAGTGAATGCAGAAACCT.. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGAGATTGTGTCCCTGGGAGAAGCAGCCt............................................................................. | 29 | t | 1.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GATGGAACTGATGTCAACCTCACCGTC.............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................TAGGGTTGCAGTGGACAGTGAATGCA......... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TAGCCCCAGATGGAACTGATGTCAACCTC.................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................ATGGAACTGATGTCAACCTC.................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCAGTGGACAGTGAATGCAGAAACCTCC | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCAGGGAATAGCCCCAGATGGAACTGATGTCAACCTCACCGTCCATGAGGCAAGCAGGGGCCACTTGTCTGTTCTTGGGGAGACGGACGTCAGGGCTCATGGTCCTCAGTGAAGAGTTACTTTAACTGGCCTGTCAGTCTCGTTGAGATTGTGTCCCTGGGAGAAGCAGCCCTGTCAACTCTGTCTGTCGTGGGTTCCCGTTCTTGTGCACTGCTAGGGTTGCAGTGGACAGTGAATGCAGAAACCTCC ...............................................................................................................................................................((((......((((((.((........)).)).)))))))).................................................. ...........................................................................................................................................140.........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................CGTGGGTTCCCGTTCTTGTGCACTGCTA................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................AGCAGCCCTGTCAAaaga....................................................................... | 18 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GTCGTGGGTTCCCGTTCTTGTGCACTGCTA................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |